isoelectric
Estimate isoelectric point for amino acid sequence
Syntax
pI = isoelectric(SeqAA)
[pI Charge] = isoelectric(SeqAA)
isoelectric(..., 'PropertyName', PropertyValue,...)
isoelectric(..., 'PKVals', PKValsValue)
isoelectric(..., 'Charge', ChargeValue)
isoelectric(..., 'Chart', ChartValue)
Arguments
SeqAA | Amino acid sequence. Enter a character vector, string, or a vector of integers from the table
Mapping Amino Acid Letter Codes to Integers. Examples: 'ARN' or
[1 2 3]. |
PKValsValue | Character vector or string specifying a file name or path and file name of a PK file
containing a table of pK values for amino acids, which .isoelectric
uses to estimate the isoelectric point (pI) of an amino acid
sequence. For an example of a PK file format, type edit Emboss.pK in the
MATLAB® command line. |
ChargeValue | Property to select a specific pH for estimating charge. Enter
a number between 0 and 14. Default
is 7.2. |
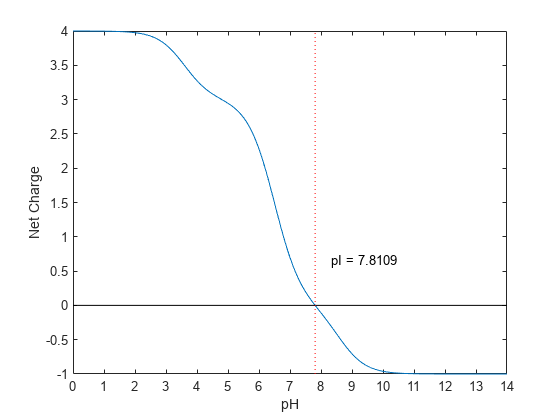
ChartValue | Controls the plotting a graph of charge versus pH. Enter true or false. |
Description
pI = isoelectric(SeqAA)pI) for
an amino acid sequence using the following pK values:
N_term 8.6 K 10.8 R 12.5 H 6.5 D 3.9 E 4.1 C 8.5 Y 10.1 C_term 3.6
The isoelectric point is the pH at which the protein has a net charge of zero.
[ returns
the estimated isoelectric point (pI Charge] = isoelectric(SeqAA)pI) for
an amino acid sequence and the estimated charge for a given pH (default
is typical intracellular pH 7.2).
The estimates are skewed by the underlying assumptions that
all amino acids are fully exposed to the solvent, that neighboring
peptides have no influence on the pK of any given amino acid, and
that the constitutive amino acids, as well as the N- and C-termini,
are unmodified. Cysteine residues participating in disulfide bridges
also affect the true pI and are not considered here. By default, isoelectric uses
the EMBOSS amino acid pK table, or you can substitute other values
using the property PKVals.
If the sequence contains ambiguous amino acid characters (b z * –),
isoelectricignores the characters and displays a warning message.Warning: Symbols other than the standard 20 amino acids appear in the sequence.
If the sequence contains undefined amino acid characters (
i j o) ,isoelectricignores the characters and displays a warning message.Warning: Sequence contains unknown characters. These will be ignored.
isoelectric(..., ' defines
optional properties using property name/value pairs.PropertyName', PropertyValue,...)
isoelectric(..., 'PKVals', uses
pK values stored in a PKValsValue)PKValValues, a PK
file, to estimate the isoelectric point (pI)
of an amino acid sequence. For an example of a PK file format, type edit
Emboss.pK in the MATLAB command line.
isoelectric(..., 'Charge', returns
the estimated charge of a sequence for a given pH (ChargeValue)ChargeValue).
isoelectric(..., 'Chart', when ChartValue)ChartValue is true,
returns a graph plotting the charge of the protein versus the pH of
the solvent.
Examples
Version History
Introduced before R2006a