gaoptimset
(Not recommended) Create genetic algorithm options structure
gaoptimset is not recommended. Use optimoptions
instead. For more information, see Version History.
Syntax
Description
gaoptimset with no input or output arguments displays a complete
list of options with their valid values. Generally, the default values appear in brackets
{}.
Note
The indicated default values are not defaults for all problem types. For an updated
list, evaluate optimoptions('ga') or
optimoptions('gamultiobj').
options = gaoptimset(Name,Value)options and sets the value of
'Name1' to Value1, 'Name2' to
Value2, and so on. gaoptimset sets any unspecified
options to [], meaning solvers use the default option values. You can
specify an option name using only enough leading characters to define the name uniquely.
gaoptimset ignores case for option names. For example,
'Display', 'display', and 'Disp'
are equivalent option names.
options = gaoptimsetoptions that contains the options for the
genetic algorithm. In this case, gaoptimset sets all option values to
[], indicating to use default values.
options = gaoptimset(@ga)options = gaoptimset(@gamultiobj)options structure containing options with explicit default values for
the ga or gamultiobj solver, respectively.
options = gaoptimset(oldopts,Name,Value)oldopts, modifying the specified options with the
specified values.
Examples
To see all the options available for ga and gamultiobj in an options structure, run gamultiobj with no input or output arguments.
gaoptimset
PopulationType: [ 'bitstring' | 'custom' | {'doubleVector'} ]
PopInitRange: [ matrix | {[-10;10]} ]
PopulationSize: [ positive scalar ]
EliteCount: [ positive scalar | {0.05*PopulationSize} ]
CrossoverFraction: [ positive scalar | {0.8} ]
ParetoFraction: [ positive scalar | {0.35} ]
MigrationDirection: [ 'both' | {'forward'} ]
MigrationInterval: [ positive scalar | {20} ]
MigrationFraction: [ positive scalar | {0.2} ]
Generations: [ positive scalar ]
TimeLimit: [ positive scalar | {Inf} ]
FitnessLimit: [ scalar | {-Inf} ]
StallGenLimit: [ positive scalar ]
StallTest: [ 'geometricWeighted' | {'averageChange'} ]
StallTimeLimit: [ positive scalar | {Inf} ]
TolFun: [ positive scalar ]
TolCon: [ positive scalar | {1e-6} ]
InitialPopulation: [ matrix | {[]} ]
InitialScores: [ column vector | {[]} ]
NonlinConAlgorithm: [ 'penalty' | {'auglag'} ]
InitialPenalty: [ positive scalar | {10} ]
PenaltyFactor: [ positive scalar | {100} ]
CreationFcn: [ function_handle | @gacreationuniform | @gacreationlinearfeasible ]
FitnessScalingFcn: [ function_handle | @fitscalingshiftlinear | @fitscalingprop |
@fitscalingtop | {@fitscalingrank} ]
SelectionFcn: [ function_handle | @selectionremainder | @selectionuniform |
@selectionroulette | @selectiontournament | @selectionstochunif ]
CrossoverFcn: [ function_handle | @crossoverheuristic | @crossoverintermediate |
@crossoversinglepoint | @crossovertwopoint | @crossoverarithmetic |
@crossoverscattered ]
MutationFcn: [ function_handle | @mutationuniform | @mutationadaptfeasible |
@mutationgaussian ]
DistanceMeasureFcn: [ function_handle | {@distancecrowding} ]
HybridFcn: [ @fminsearch | @patternsearch | @fminunc | @fmincon | {[]} ]
Display: [ 'off' | 'iter' | 'diagnose' | {'final'} ]
OutputFcns: [ function_handle | {[]} ]
PlotFcns: [ function_handle | @gaplotbestf | @gaplotbestindiv | @gaplotdistance |
@gaplotexpectation | @gaplotgenealogy | @gaplotselection | @gaplotrange |
@gaplotscorediversity | @gaplotscores | @gaplotstopping |
@gaplotmaxconstr | @gaplotrankhist | @gaplotpareto | @gaplotspread |
@gaplotparetodistance |{[]} ]
PlotInterval: [ positive scalar | {1} ]
Vectorized: [ 'on' | {'off'} ]
UseParallel: [ logical scalar | true | {false} ]
Set options for ga to have a starting population that includes the point [1,1] and to use the gaplotbestf plot function.
options = gaoptimset('InitialPopulation',[1 1],... 'PlotFcns',@gaplotbestf);
Find a local minimum of the function rastriginsfcn, which is available when you run this example, using the specified options.
rng default % For reproducibility nvar = 2; A = []; b = []; Aeq = []; beq = []; lb = []; ub = []; nlconst = []; [x,fval] = ga(@rastriginsfcn,nvar,A,b,Aeq,beq,lb,ub,nlconst,options)
ga stopped because the average change in the fitness value is less than options.FunctionTolerance.
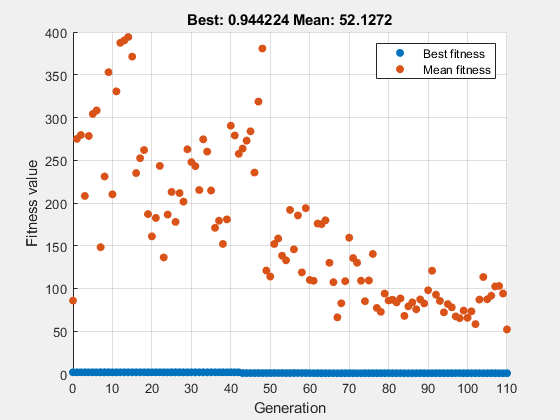
x = 1×2
0.0227 0.0656
fval = 0.9442
Use gaoptimset to create an option structure for ga with the values set to their defaults.
options = gaoptimset(@ga)
options = struct with fields:
PopulationType: 'doubleVector'
PopInitRange: []
PopulationSize: 'auto'
EliteCount: '0.05*PopulationSize'
CrossoverFraction: 0.8000
ParetoFraction: []
MigrationDirection: 'forward'
MigrationInterval: 20
MigrationFraction: 0.2000
Generations: '100*numberOfVariables'
TimeLimit: Inf
FitnessLimit: -Inf
StallGenLimit: 50
StallTest: 'averageChange'
StallTimeLimit: Inf
TolFun: 1.0000e-06
TolCon: 1.0000e-03
InitialPopulation: []
InitialScores: []
NonlinConAlgorithm: 'auglag'
InitialPenalty: 10
PenaltyFactor: 100
PlotInterval: 1
CreationFcn: []
FitnessScalingFcn: @fitscalingrank
SelectionFcn: []
CrossoverFcn: []
MutationFcn: []
DistanceMeasureFcn: []
HybridFcn: []
Display: 'final'
PlotFcns: []
OutputFcns: []
Vectorized: 'off'
IntegerTolerance: 1.0000e-05
UseParallel: 0
Create genetic algorithm options to return an iterative display.
oldopts = gaoptimset('Display','iter');
Modify oldopts to include the gaplotbestf plot function and a population size of 250.
options = gaoptimset(oldopts,'PlotFcns',@gaplotbestf,... 'PopulationSize',250)
options = struct with fields:
PopulationType: []
PopInitRange: []
PopulationSize: 250
EliteCount: []
CrossoverFraction: []
ParetoFraction: []
MigrationDirection: []
MigrationInterval: []
MigrationFraction: []
Generations: []
TimeLimit: []
FitnessLimit: []
StallGenLimit: []
StallTest: []
StallTimeLimit: []
TolFun: []
TolCon: []
InitialPopulation: []
InitialScores: []
NonlinConAlgorithm: []
InitialPenalty: []
PenaltyFactor: []
PlotInterval: []
CreationFcn: []
FitnessScalingFcn: []
SelectionFcn: []
CrossoverFcn: []
MutationFcn: []
DistanceMeasureFcn: []
HybridFcn: []
Display: 'iter'
PlotFcns: @gaplotbestf
OutputFcns: []
Vectorized: []
IntegerTolerance: []
UseParallel: []
Create two sets of genetic algorithm options, oldopts and newopts. Specify an iterative display and the gaplotbestf plot function for oldopts. Specify no display and a population size of 300 for newopts.
oldopts = gaoptimset('Display','iter','PlotFcns',@gaplotbestf); newopts = gaoptimset('Display','off','PopulationSize',300);
Combine these options with newopts taking precedence.
options = gaoptimset(oldopts,newopts)
options = struct with fields:
PopulationType: []
PopInitRange: []
PopulationSize: 300
EliteCount: []
CrossoverFraction: []
ParetoFraction: []
MigrationDirection: []
MigrationInterval: []
MigrationFraction: []
Generations: []
TimeLimit: []
FitnessLimit: []
StallGenLimit: []
StallTest: []
StallTimeLimit: []
TolFun: []
TolCon: []
InitialPopulation: []
InitialScores: []
NonlinConAlgorithm: []
InitialPenalty: []
PenaltyFactor: []
PlotInterval: []
CreationFcn: []
FitnessScalingFcn: []
SelectionFcn: []
CrossoverFcn: []
MutationFcn: []
DistanceMeasureFcn: []
HybridFcn: []
Display: 'off'
PlotFcns: @gaplotbestf
OutputFcns: []
Vectorized: []
IntegerTolerance: []
UseParallel: []
Notice that the value of the Display option is the value specified in newopts.
Input Arguments
Optimization options, specified as a structure, such as the output of
gaoptimset.
Data Types: struct
Optimization options, specified as a structure, such as the output of
gaoptimset. Any options in newopts with
nonempty values overwrite the corresponding options in oldopts in
this syntax:
options = gaoptimset(oldopts,newopts)
Data Types: struct
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: options =
gaoptimset('Display','off','PlotFcns',@gaplotbestf)
gaoptimset creates a structure. In the following table of option
names, use the name at the bottom of the description after the phrase "For an options
structure," if specified. The first listed name (Option) is for the
optimoptions function, which is the recommended function for setting
options. For example, to specify the tolerance on nonlinear constraint violation in
optimoptions you set 'ConstraintTolerance', but
for gaoptimset you set 'TolCon'.
In the following table,
Values in braces
{}denote the default value.{}*represents the default when the problem has linear constraints, and whenMutationFcnhas bounds.I* indicates that
gahandles options for integer constraints differently; this notation does not apply togamultiobj.NM indicates that the option does not apply to
gamultiobj.
optimoptions hides the options listed in
italics; see Options that optimoptions Hides.
Options for ga and gamultiobj
| Option | Description | Values | |
|---|---|---|---|
ConstraintTolerance | Determines the feasibility with respect
to nonlinear constraints. Also,
For an options structure, use
| Nonnegative scalar | |
| Function that creates the initial population. Specify as a name of a built-in creation function or a function handle. See Population Options. Defaults: | Choose any of these:
| |
Integer:
Else nonlinear
with
Else
linear: Otherwise:
|
Integer:
Else linear:
Otherwise:
| ||
| Function that the algorithm uses to create crossover children. Specify as a name of a built-in crossover function or a function handle. See Crossover Options. Defaults: | Choose any of these:
| |
Integer:
Else linear:
Otherwise:
|
Integer
or linear: Otherwise:
| ||
| The fraction of the population at the
next generation, not including elite children, that the crossover function
creates. Default: | Nonnegative scalar | |
| Level of display. Default:
|
| |
| Function that computes the distance
measure of individuals. Specify as a name of a built-in distance measure
function or a function handle. The value applies to the decision variable or
design space (genotype) or to function space (phenotype). The default
For an options structure, use a function handle, not a name. |
| |
| Positive integer specifying how many
individuals in the current generation are guaranteed to survive to the next
generation. Not used in | Nonnegative integer | |
| If the fitness function attains the
value of | Scalar | |
| Function that scales the values of the
fitness function. Specify as a name of a built-in scaling function or a
function handle. Not used in |
| |
FunctionTolerance | The algorithm stops if the average
relative change in the best fitness function value over
For
For an options structure, use
| Nonnegative scalar | | |
| Function that continues the
optimization after Alternatively, a cell array specifying the hybrid function and its options. See ga Hybrid Function. For When the problem has integer constraints, you cannot use a hybrid function. | Function name or handle | or 1-by-2
cell array | | |
InitialPenalty | Initial value of the penalty parameter.
Not used in | Positive scalar | |
| Initial population used to seed the
genetic algorithm. Has up to For an options structure, use
| Matrix | |
| Matrix or vector specifying the range
of the individuals in the initial population. Applies to
For an options structure, use
| Matrix or vector | |
| Initial scores used to determine
fitness. Has up to For an options structure, use
| Column vector for single objective | matrix for multiobjective | |
| Maximum number of iterations before the
algorithm halts. Default: For an options structure,
use | Nonnegative integer | |
| The algorithm stops if the average
relative change in the best fitness function value over
For
For an options structure, use
| Nonnegative integer | |
| The algorithm stops if there is no
improvement in the objective function for For an options structure, use
| Positive scalar | |
| The algorithm stops after running for
For an
options structure, use | Nonnegative scalar | |
MigrationDirection | Direction of migration. Default:
|
| |
MigrationFraction | Scalar from 0 through 1 specifying the
fraction of individuals in each subpopulation that migrates to a different
subpopulation. Default: | Scalar | |
MigrationInterval | Positive integer specifying the number
of generations that take place between migrations of individuals between
subpopulations. Default: | Positive integer | |
| Function that produces mutation children. Specify as a name of a built-in mutation function or a function handle. See Mutation Options. Defaults: | Choose any of these:
| |
Integer:
Else linear or bounds:
Otherwise:
|
Integer:
Else linear or
bounds: Otherwise:
| ||
| Nonlinear constraint algorithm. See
Nonlinear Constraint Solver Algorithms for Genetic Algorithm.
Option unchangeable for For an options structure, use
|
| |
| Functions that the solver calls at each
iteration. Specify as a function handle or a cell array of function handles.
Default: For an options structure,
use | Function handle or cell array of function handles | |
| Scalar from 0 through 1 specifying the
fraction of individuals to keep on the first Pareto front while the solver
selects individuals from higher fronts, for | Scalar | | |
PenaltyFactor | Penalty update parameter. Not used in
| Positive scalar | |
| Function that plots data computed by the algorithm. Specify as a name of a built-in plot function, a function handle, or a cell array of built-in names or function handles. See Plot Options. For an options structure, use
|
| |
PlotInterval | Positive integer specifying the number
of generations between consecutive calls to the plot functions. Default:
| Positive integer | |
| Size of the population. Default:
| Positive integer | |
| Data type of the population. Must be
|
| |
| Function that selects parents of crossover and mutation children. Specify as a name of a built-in selection function or a function handle. Defaults: | For
| |
Integer or
nonlinear penalty:
Otherwise:
|
| ||
StallTest | Stopping test type. Not used in
|
| |
UseParallel | Compute fitness and nonlinear
constraint functions in parallel. Default: |
| |
| Specifies whether functions are
vectorized. Default: For an options structure, use |
| |
Output Arguments
Optimization options, returned as a structure.
Version History
Introduced before R2006aTo set options, the gaoptimset, psoptimset, and
saoptimset functions are not recommended. Instead, use optimoptions.
The main difference between using optimoptions and the other
functions is that you include the solver name as the first argument in
optimoptions. For example, to set an iterative display in
ga:
options = optimoptions('ga','Display','iter'); % instead of options = gaoptimset('Display','iter');
The other difference is that some option names have changed. You can continue to use the
old names in optimoptions. For details, see Options Changes in R2016a.
optimoptions offers these advantages over the other functions:
optimoptionsprovides better automatic code suggestions and completions, especially in the Live Editor.You can use a single option-setting function instead of a variety of functions.
There are no plans to remove gaoptimset,
psoptimset, and saoptimset at this
time.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)