imkeepborder
Description
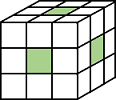
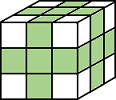
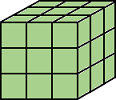
J = imkeepborder(I)I that are lighter
than their surroundings and connected to the image border, while suppressing all
other structures. If I is a binary image, then a structure is a connected group of white pixels. Use
this function to keep the image border while clearing structures that are not
touching the border. The output image J is grayscale or binary,
depending on the input.
J = imkeepborder(I,Name=Value)imkeepborder(I,Borders=["left" "right"])
retains only the structures touching the left or right image border.
Examples
Input Arguments
Output Arguments
Algorithms
imkeepborder uses morphological reconstruction where:
The mask image is the input image.
The marker image is 0 everywhere except along the border, where it equals the mask image.
References
[1] Soille, Pierre. Morphological Image Analysis: Principles and Applications Berlin ; New York: Springer, 1999, 164–165.
[2] Molnar, Ian. Uniform quartz - Silver nanoparticle injection experiment, Digital Rocks Portal (April 2016). Accessed March 10, 2023. https://www.digitalrocksportal.org/projects/44, made available under the ODC-BY 1.0 Attribution License.
Extended Capabilities
Version History
Introduced in R2023b