Train Custom Cellpose Model
This example shows how to train a custom Cellpose model, using new training data, to detect noncircular shapes.
The Cellpose Library provides several pretrained models that have been trained to detect cells in microscopy images. The pretrained models can often detect other circular objects. For example, you can use the pretrained cyto2 model to label pears or coins by specifying the segmentCells2D function with an ImageCellDiameter value that reflects the approximate object diameter, in pixels. To detect other shapes, you can train a custom Cellpose model using training images with the target shape. In this example, you train a custom model that detects diamond shapes.

This example requires the Medical Imaging Toolbox™ Interface for Cellpose Library. You can install the Medical Imaging Toolbox Interface for Cellpose Library from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons. The Medical Imaging Toolbox Interface for Cellpose Library requires Deep Learning Toolbox™ and Computer Vision Toolbox™.
Create Training and Testing Images
Create a new image data set to train a custom model. For this example, create a data set to train a model that detects diamond-shaped objects.
First, create a structuring element corresponding to the object shape and size you want to detect. For this example, create a diamond in which the center and corner points are 35 pixels apart.
radius = 35;
sd = strel("diamond",radius);Specify the target size of the images, in pixels, and the number of diamonds to include in each image.
imageSize = [128 128]; numObjectsPerImage = 1;
Create training images by using the makeRepeatedShapeData helper function, which is defined at the end of this example. Save the test image to a subfolder named train within the current directory.
numTrainingImages = 15; trainingFolderName = "train"; makeRepeatedShapeData(trainingFolderName,sd, ... numTrainingImages,imageSize,numObjectsPerImage)
Create one test image using the makeRepeatedShapeData helper function. Save the test image to a subfolder named test within the current directory.
numTestingImages = 1; testFolderName = "test"; makeRepeatedShapeData(testFolderName,sd, ... numTestingImages,imageSize,numObjectsPerImage)
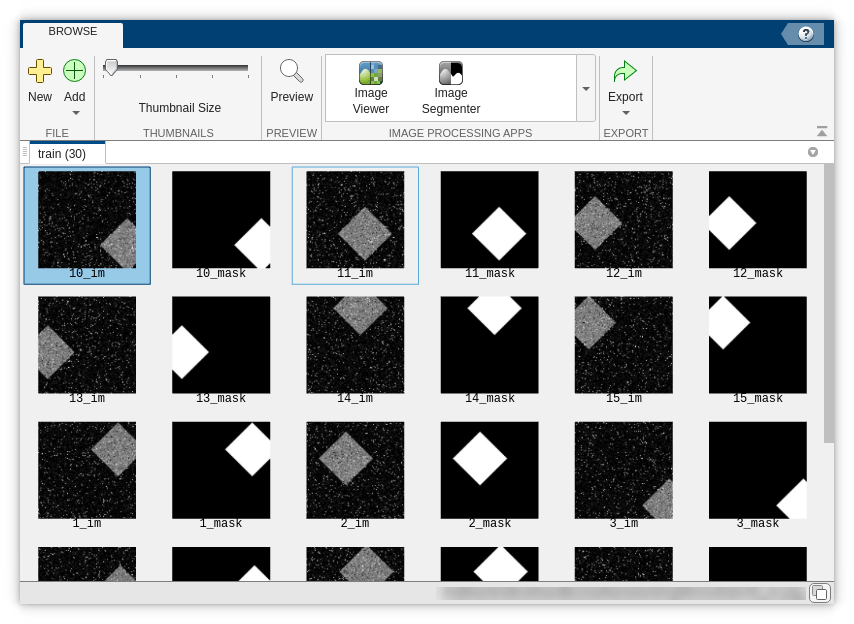
You can visually check the training and test images by using the Image Browser app.
imageBrowser(trainingFolderName)
Test Pretrained Model
Test whether the pretrained cyto2 model can detect the diamonds in the test image.
cp = cellpose(Model="cyto2"); imTest = imread(testFolderName + filesep + "1_im.png"); labelPretrained = segmentCells2D(cp,imTest,ImageCellDiameter=2*radius);
Display the test image and the labels predicted by the pretrained cyto2 model. The label mask is empty, indicating that the model does not detect the diamond.
figure tiledlayout(1,2,TileSpacing="tight") nexttile imshow(imTest) title("Test Image") nexttile imshow(labeloverlay(imTest,labelPretrained)) title("Label from Pretrained cyto2")

Train Custom Model
Train a custom Cellpose model by using the trainCellpose function. Specify the training data location and the name for the new model as the first two input arguments. Use name-value arguments to specify additional training details. The PretrainedModel name-value argument specifies whether to start training from one of the pretrained models in the Cellpose library. This example retrains a copy of the pretrained cyto2 model. This is the default choice, and is generally suitable unless your training data is similar to that of another pretrained model. To learn more about each model and its training data, see the Models page of the Cellpose Library Documentation. Specify the ImageSuffix and LabelSuffix arguments to match the filenames of the images and masks in the training data set, respectively. The function saves the new model in the current directory.
By default, the function trains on a GPU if one is available, and otherwise trains on the CPU. Training on a GPU requires Parallel Computing Toolbox™. For information on supported devices, see GPU Computing Requirements (Parallel Computing Toolbox).
OutputModelFile = "diamondDetectionModel"; trainCellpose(trainingFolderName,OutputModelFile,... PreTrainedModel="cyto2", ... MaxEpoch=2, ... ImageSuffix="_im", ... LabelSuffix="_mask");
INFO:cellpose.io:30 / 30 images in C:\train folder have labels INFO:cellpose.models:>> cyto2 << model set to be used INFO:cellpose.models:>>>> model diam_mean = 30.000 (ROIs rescaled to this size during training) INFO:cellpose.dynamics:flows precomputed INFO:cellpose.core:>>>> median diameter set to = 30 INFO:cellpose.core:>>>> mean of training label mask diameters (saved to model) 52.052 INFO:cellpose.core:>>>> training network with 2 channel input <<<< INFO:cellpose.core:>>>> LR: 0.20000, batch_size: 8, weight_decay: 0.00001 INFO:cellpose.core:>>>> ntrain = 30 INFO:cellpose.core:>>>> nimg_per_epoch = 30 INFO:cellpose.core:Epoch 0, Time 0.9s, Loss 0.3013, LR 0.0000 INFO:cellpose.core:saving network parameters to C:\models/cellpose_residual_on_style_on_concatenation_off__2023_07_21_08_55_07.570863_epoch_1
Test Custom Model
Examine the custom model by using it to create a new cellpose object. The TrainingCellDiameter property value now reflects the size of the diamonds in the training image masks.
cpt = cellpose(Model=OutputModelFile); cpt.TrainingCellDiameter
ans = 52.0520
Test the custom model by segmenting the test image.
labelCustom = segmentCells2D(cpt,imTest);
Display the result. The custom model correctly labels the diamond shape.
figure tiledlayout(1,2,TileSpacing="tight") nexttile imshow(imTest) title("Test Image") nexttile imshow(labeloverlay(imTest,labelCustom)) title("Label from Custom Model")

Helper Functions
The makeRepeatedShapeData helper function creates images and label masks by performing these steps:
Creates images of the size specified by
imageSizethat contain the shape described by a structuring element,sd, plus salt and pepper noise.Creates a binary ground truth label mask showing the location of the shape.
Saves the images and ground truth masks as PNG files to a subfolder named
trainwithin the current directory. If the subfolder does not exist, the function creates it before saving the images.
function makeRepeatedShapeData(folderName,sd,numImages,imageSize,numObjectsPerImage) % Set the random number seed to generate consistent images across runs rng(0) % Create the specified folder if it does not exist if ~exist(folderName,"dir") mkdir(folderName); end % Convert the structuring element to a numeric matrix shape = double(sd.Neighborhood)*127; % Create and save the images for ind = 1:numImages img = zeros(imageSize,"uint8"); % Seed the shape centers objCenters = randi(numel(img),numObjectsPerImage,1); img(objCenters) = 1; % "Stamp" the shapes into the image img = imfilter(img,shape); % Create the mask mask = img > 0; % Add noise to the image img = imnoise(img); img = imnoise(img,"salt & pepper"); % Save the image and mask baseFileName = folderName + filesep + string(ind); imwrite(img,baseFileName + "_im.png"); imwrite(mask,baseFileName + "_mask.png"); end end
References
[1] Stringer, Carsen, Tim Wang, Michalis Michaelos, and Marius Pachitariu. “Cellpose: A Generalist Algorithm for Cellular Segmentation.” Nature Methods 18, no. 1 (January 2021): 100–106. https://doi.org/10.1038/s41592-020-01018-x.
[2] Pachitariu, Marius, and Carsen Stringer. “Cellpose 2.0: How to Train Your Own Model.” Nature Methods 19, no. 12 (December 2022): 1634–41. https://doi.org/10.1038/s41592-022-01663-4.
See Also
cellpose (Medical Imaging Toolbox) | segmentCells2D (Medical Imaging Toolbox) | segmentCells3D (Medical Imaging Toolbox) | trainCellpose (Medical Imaging Toolbox) | downloadCellposeModels (Medical Imaging Toolbox)
Topics
- Choose Pretrained Cellpose Model for Cell Segmentation
- Refine Cellpose Segmentation by Tuning Model Parameters
- Detect Nuclei in Large Whole Slide Images Using Cellpose