Results for
In 2019, I wrote a MATLAB Central blog post called "The tool builder's gene (or how to get a job at MathWorks)." In it, I explained my personal theory of a characteristic of some engineers that is key for becoming successful software developers at MathWorks.
I just shared this essay on my personal blog, along with a couple of updates.
What is MATLAB Project?
40%
Never use it
28%
Only use existing from others' proj
3%
Use it occasionally
13%
Use it frequently
16%
90 votes
clc; clear; close all;
% Initial guess for [x1, x2, x3] (adjust as needed)
x0 = [0.2,0.35,0.5];
% No linear constraints
A = []; b = [];
Aeq = []; beq = [];
% Lower and upper bounds (adjust based on the problem)
lb = [0,0,0];
ub = [pi/2,pi/2,pi/2];
% Optimization options
options = optimoptions('fmincon', 'Algorithm', 'sqp', 'Display', 'iter');
% Solve with fmincon
[x_opt, fval, exitflag] = fmincon(@objective, x0, A, b, Aeq, beq, lb, ub, @nonlinear_constraints, options);
% Display results
fprintf('Optimal Solution: x1 = %.4f, x2 = %.4f, x3 = %.4f\n', x_opt(1), x_opt(2), x_opt(3));
fprintf('Exit Flag: %d\n', exitflag);
%% Objective function (minimizing sum of squared errors)
function f = objective(x)
f = sum(x.^2); % Dummy function (since we only want to solve equations)
end
%% Nonlinear constraints (representing the trigonometric equations)
function [c, ceq] = nonlinear_constraints(x)
% Example nonlinear trigonometric equations:
ceq(1) = cos(x(1))+cos(x(2))+cos(x(3))-3*0.9; % First equation
ceq(2) = cos(5*x(1))+cos(5*x(2))+cos(5*x(3)); % Second equation
ceq(3) = cos(7*x(1))+cos(7*x(2))+cos(7*x(3)); % Third equation
c = [x(1)-x(2); x(2)-x(3)]; % No inequality constraints
end
Can anyone provide some matlab learning paths, I am a novice to MATLAB, I would appreciate it
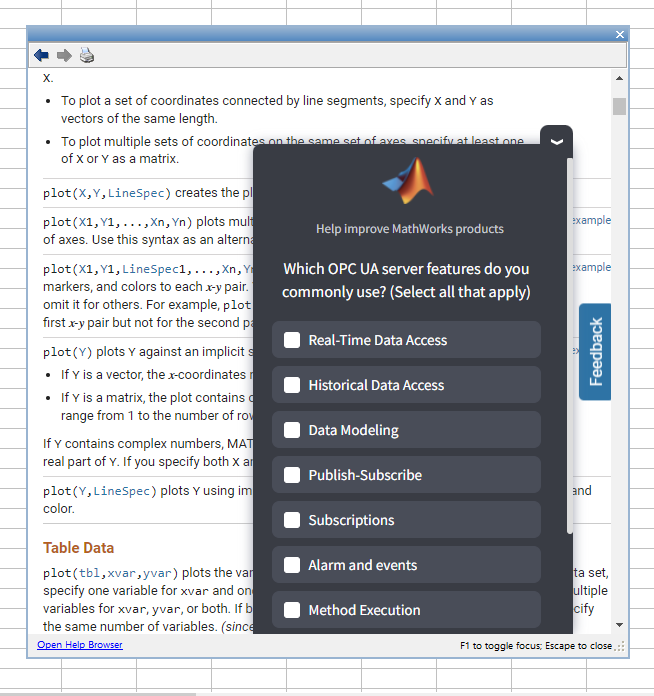
I'm getting this annoying survey (screenshot below) in the help windows of MATLAB R2024b this morning. It blocks the text I'm actually trying to read, when minimised it pops up again after a few minutes, and persists even after picking an option and completing the SurveyMonkey survey it links to. I don't even know what the OPC UA server so rest assured any of my answers to that survey aren't going to help MathWorks improve their product.
It is April 3, 2025 now. Where is the MATLAB 2025a?
Me: If you have parallel code and you apply this trick that only requires changing one line then it might go faster.
Reddit user: I did and it made my code 3x faster
Not bad for just one line of code!
Which makes me wonder. Could it make your MATLAB program go faster too? If you have some MATLAB code that makes use of parallel constructs like parfor or parfeval then start up your parallel pool like this
parpool("Threads")
before running your program.
The worst that will happen is you get an error message and you'll send us a bug report....or maybe it doesn't speed up much at all....
....or maybe you'll be like the Reddit user and get 3x speed-up for 10 seconds work. It must be worth a try...after all, you're using parallel computing to make your code faster right? May as well go all the way.
In an artificial benchmark I tried, I got 10x speedup! More details in my recent blog post: Parallel computing in MATLAB: Have you tried ThreadPools yet? » The MATLAB Blog - MATLAB & Simulink
Give it a try and let me know how you get on.
I am pleased to announce the 6th Edition of my book MATLAB Recipes for Earth Sciences with Springer Nature
also in the MathWorks Book Program
It is now almost exactly 20 years since I signed the contract with Springer for the first edition of the book. Since then, the book has grown from 237 to 576 pages, with many new chapters added. I would like to thank my colleagues Norbert Marwan and Robin Gebbers, who have each contributed two sections to Chapters 5, 7 and 9.
And of course, my thanks go to the excellent team at the MathWorks Book Program and the numerous other MathWorks experts who have helped and advised me during the last 30+ years working with MATLAB. And of course, thank you Springer for 20 years of support.
This book introduces methods of data analysis in the earth sciences using MATLAB, such as basic statistics for univariate, bivariate, and multivariate data sets, time series analysis, signal processing, spatial and directional data analysis, and image analysis.
Martin H. Trauth

There has been a lot of discussion here about the R2025a Prerelease that has really helped us get it ready for the prime time. Thank you for that!
A new update of the Prerelease has just dropped. So fresh it is still warm from the oven! In my latest blog post I discuss changes in the way MathWorks has been asking-for and processing feedback...and you have all been a part of that.
If you haven't tried the Prerelease in a while, I suggest you update and see how things are looking now.
If you have already submitted a bug report and it hasn't been fixed in this update, you don't need to submit another one. Everything is being tracked!
Have a play, discuss it here and thanks for again for being part of the process.
📢 We want to hear from you! We're a team of graduate student researchers at the University of Michigan studying MATLAB Drive and other cloud-based systems for sharing coding files. Your feedback will help improve these tools. Take our quick survey here: https://forms.gle/DnHs4XNAwBZvmrAw6
No
50%
Yes, but I am not interested
8%
Yes, but it is too expensive
20%
Yes, I would like to know more
18%
Yes, I am cert. MATLAB Associate
2%
Yes, I am cert. MATLAB Professional
3%
4779 votes
I am glad to inform and share with you all my new text book titled "Inverters and AC Drives
Control, Modeling, and Simulation Using Simulink", Springer, 2024. This text book has nine chapters and three appendices. A separate "Instructor Manual" is rpovided with solutions to selected model projects. The salent features of this book are given below:
- Provides Simulink models for various PWM techniques used for inverters
- Presents vector and direct torque control of inverter-fed AC drives and fuzzy logic control of converter-fed AC drives
- Includes examples, case studies, source codes of models, and model projects from all the chapters
The Springer link for this text book is given below:
This book is also in the Mathworks book program:
Over the last 5 years or so, the highest-traffic post on my MATLAB Central image processing blog was not actually about image processing; it was about changing the default line thickness in plots.
Now I have written about some other MATLAB plotting behavior that I have recently changed to suit my own preferences. See this new blog post.
Here is a standard MATLAB plot:
x = 0:pi/100:2*pi;
y1 = sin(x);
y2 = cos(x);
plot(x,y1,x,y2)
I don't like some aspects of this plot, and so I have put the following code into my startup file.
set(groot,"DefaultLineLineWidth",2)
set(groot,"DefaultAxesXLimitMethod","padded")
set(groot,"DefaultAxesYLimitMethod","padded")
set(groot,"DefaultAxesZLimitMethod","padded")
set(groot,"DefaultAxesXGrid","on")
set(groot,"DefaultAxesYGrid","on")
set(groot,"DefaultAxesZGrid","on")
With those defaults changed, here is my preferred appearance:
plot(x,y1,x,y2)
To develop uifigure-based app, I wish MATLAB can provide something like uiquestdlg to replace questdlg without changing too much of the original code developed for figure-based app. Also, uiinputdlg <-> inputdlg and so on.
Learn the basic of quantum computing, how to simulate quantum circuits on MATLAB and how to run them on real quantum computers using Amazon Braket. There will also be a demonstration of machine learning using quantum computers!

Details at MATLAB-AMAZON Braket Hands-on Quantum Machine Learning Workshop - MATLAB & Simulink. This will be led by MathWorker Hossein Jooya.
I kicked off my own exploration of Quantum Computing in MATLAB a year or so ago and wrote it up on The MATLAB Blog: Quantum computing in MATLAB R2023b: On the desktop and in the cloud » The MATLAB Blog - MATLAB & Simulink. This made use of the MATLAB Support Package for Quantum Computing - File Exchange - MATLAB Central
Good day I am looking someone to help me on the matlab and simulink I am missing some explanations.For easy communication you can contact 0026876637042
Recently my iMac became sluggish. I checked Activity Monitor and found it was spending most of its time in mds_stores. I turned of Apple Intelligence under System Settings - Apple Intelligence & Siri, and its like new again.
The GCD approach to identify rough numbers is a terribly useful one, well worth remembering. But at some point, I expect someone to notice that all work done with these massively large symbolic numbers uses only one of the cores on your computer. And, having spent so much money on those extra cores in your CPU, surely we can find a way to use them all? The problem is, computations done on symbolic integers never use more than 1 core. (Sad, unhappy face.)
In order to use all of the power available to your computer using MATLAB, you need to work in double precision, or perhaps int64 or uint64. To do that, I'll next search for primes among the family 3^n+4. In fact, they seem pretty common, at least if we look at the first few such examples.
F = @(n) sym(3).^n + 4;
F(0:16)
ans =
[5, 7, 13, 31, 85, 247, 733, 2191, 6565, 19687, 59053, 177151, 531445, 1594327, 4782973, 14348911, 43046725]
isprime(F(0:16))
ans =
1×17 logical array
1 1 1 1 0 0 1 0 0 1 1 0 0 0 0 0 0
Of the first 11 members of that sequence, 7 of them were prime. Naturally, primes will become less frequent in this sequence as we look further out. The members of this family grow rapidly in size. F(10000) has 4771 decimal digits, and F(100000) has 47712 decimal digits. We certainly don't want to directly test every member of that sequence for primality. However, what I will call a partial or incomplete sieve can greatly decrease the work needed.
Consider there are roughly 5.7 million primes less than 1e8.
numel(primes(1e8))
ans =
5761455
F(17) is the first member of our sequence that exceeds 1e8. So we can start there, since we already know the small-ish primes in this sequence.
roughlim = 1e8;
primes1e8 = primes(roughlim);
primes1e8([1 2]) = []; % F(n) is never divisible by 2 or 3
F_17 = double(F(17));
Fremainders = mod(F_17,primes1e8);
nmax = 100000;
FnIsRough = false(1,nmax);
for n = 17:nmax
if all(Fremainders)
FnIsRough(n) = true;
end
% update the remainders for the next term in the sequence
% This uses the recursion: F(n+1) = 3*F(n) - 8
Fremainders = mod(Fremainders*3 - 8,primes1e8);
end
sum(FnIsRough)
ans =
6876
These will be effectively trial divides, even though we use mod for the purpose. The result is 6876 1e8-rough numbers, far less than that total set of 99984 values for n. One thing of great importance is to recognize this sequence of tests will use an approximately constant time per test regardless of the size of the numbers because each test works off the remainders from the previous one. And that works as long as we can update those remainders in some simple, direct, and efficient fashion. All that matters is the size of the set of primes to test against. Remember, the beauty of this scheme is that while I did what are implicitly trial divides against 5.76 million primes at each step, ALL of the work was done in double precision. That means I used all 8 of the cores on my computer, pushing them as hard as I could. I never had to go into the realm of big integer arithmetic to identify the rough members in that sequence, and by staying in the realm of doubles, MATLAB will automatically use all the cores you have available.
The first 10 values of n (where n is at least 17), such that F(n) is 1e8-rough were
FnIsRough = find(FnIsRough);
FnIsRough(1:10)
ans =
22 30 42 57 87 94 166 174 195 198
How well does the roughness test do to eliminate composite members of this sequence?
isprime(F(FnIsRough(1:10)))
ans =
1×10 logical array
1 1 1 1 1 0 0 1 1 1
As you can see, 8 of those first few 1e8-rough members were actually prime, so only 2 of those eventual isprime tests were effectively wasted. That means the roughness test was quite useful indeed as an efficient but relatively weak pre-test for possible primality. More importantly it is a way to quickly eliminate those values which can be known to be composite.
You can apply a similar set of tests on many families of numbers. For example, repunit primes are a great case. A rep-digit number is any number composed of a sequence of only a single digit, like 11, 777, and 9999999999999.
However, you should understand that only rep-digit numbers composed of entirely ones can ever be prime. Naturally, any number composed entirely of the digit D, will always be divisible by the single digit number D, and so only rep-unit numbers can be prime. Repunit numbers are a subset of the rep-digit family, so numbers composed only of a string of ones. 11 is the first such repunit prime. We can write them in MATLAB as a simple expression:
RU = @(N) (sym(10).^N - 1)/9;
RU(N) is a number composed only of the digit 1, with N decimal digits. This family also follows a recurrence relation, and so we could use a similar scheme as was used to find rough members of the set 3^N-4.
RU(N+1) == 10*RU(N) + 1
However, repunit numbers are rarely prime. Looking out as far as 500 digit repunit numbers, we would see primes are pretty scarce in this specific family.
find(isprime(RU(1:500)))
ans =
2 19 23 317
There are of course good reasons why repunit numbers are rarely prime. One of them is they can only ever be prime when the number of digits is also prime. This is easy to show, as you can always factor any repunit number with a composite number of digits in a simple way:
1111 (4 digits) = 11*101
111111111 (9 digits) = 111*1001001
Finally, I'll mention that Mersenne primes are indeed another example of repunit primes, when expressed in base 2. A fun fact: a Mersenne number of the form 2^n-1, when n is prime, can only have prime factors of the form 1+2*k*n. Even the Mersenne number itself will be of the same general form. And remember that a Mersenne number M(n) can only ever be prime when n is itself prime. Try it! For example, 11 is prime.
Mn = @(n) sym(2).^n - 1;
Mn(11)
ans =
2047
Note that 2047 = 1 + 186*11. But M(11) is not itself prime.
factor(Mn(11))
ans =
[23, 89]
Looking carefully at both of those factors, we see that 23 == 1+2*11, and 89 = 1+8*11.
How does this help us? Perhaps you may see where this is going. The largest known Mersenne prime at this date is Mn(136279841). This is one seriously massive prime, containing 41,024,320 decimal digits. I have no plans to directly test numbers of that size for primality here, at least not with my current computing capacity. Regardless, even at that realm of immensity, we can still do something.
If the largest known Mersenne prime comes from n=136279841, then the next such prime must have a larger prime exponent. What are the next few primes that exceed 136279841?
np = NaN(1,11); np(1) = 136279841;
for i = 1:10
np(i+1) = nextprime(np(i)+1);
end
np(1) = [];
np
np =
Columns 1 through 8
136279879 136279901 136279919 136279933 136279967 136279981 136279987 136280003
Columns 9 through 10
136280009 136280051
The next 10 candidates for Mersenne primality lie in the set Mn(np), though it is unlikely that any of those Mersenne numbers will be prime. But ... is it possible that any of them may form the next Mersenne prime? At the very least, we can exclude a few of them.
for i = 1:10
2*find(powermod(sym(2),np(i),1+2*(1:50000)*np(i))==1)
end
ans =
18 40 64
ans =
1×0 empty double row vector
ans =
2
ans =
1×0 empty double row vector
ans =
1×0 empty double row vector
ans =
1×0 empty double row vector
ans =
1×0 empty double row vector
ans =
1×0 empty double row vector
ans =
1×0 empty double row vector
ans =
2
Even with this quick test which took only a few seconds to run on my computer, we see that 3 of those Mersenne numbers are clearly not prime. In fact, we already know three of the factors of M(136279879), as 1+[18,40,64]*136279879.
You might ask, when is the MOD style test, using a large scale test for roughness against many thousands or millions of small primes, when is it better than the use of GCD? The answer here is clear. Use the large scale mod test when you can easily move from one member of the family to the next, typically using a linear recurrence. Simple such examples of this are:
1. Repunit numbers
General form: R(n) = (10^n-1)/9
Recurrence: R(n+1) = 10*R(n) + 1, R(0) = 1, R(1) = 11
2. Fibonacci numbers.
Recurrence: F(n+1) = F(n) + F(n-1), F(0) = 0, F(1) = 1
3. Mersenne numbers.
General form: M(n) = 2^n - 1
Recurrence: M(n+1) = 2*M(n) + 1
4. Cullen numbers, https://en.wikipedia.org/wiki/Cullen_number
General form: C(n) = n*2^n + 1
Recurrence: C(n+1) = 4*C(n) + 4*C(n-1) + 1
5. Hampshire numbers: (My own choice of name)
General form: H(n,b) = (n+1)*b^n - 1
Recurrence: H(n+1,b) = 2*b*H(n-1,b) - b^2*H(n-2,b) - (b-1)^2, H(0,b) = 0, H(1,b) = 2*b-1
6. Tin numbers, so named because Sn is the atomic symbol for tin.
General form: S(n) = 2*n*F(n) + 1, where F(n) is the nth Fibonacci number.
Recurrence: S(n) = S(n-5) + S(n-4) - 3*S(n-3) - S(n-2) +3*S(n-1);
To wrap thing up, I hope you have enjoyed this beginning of a journey into large primes and non-primes. I've shown a few ways we can use roughness, first in a constructive way to identify numbers which may harbor primes in a greater density than would otherwise be expected. Next, using GCD in a very pretty way, and finally by use of MOD and the full power of MATLAB to test elements of a sequence of numbers for potential primality.
My next post will delve into the world of Fermat and his little theorem, showing how it can be used as a stronger test for primality (though not perfect.)
Yes, some readers might now argue that I used roughness in a crazy way in my last post, in my approach to finding a large twin prime pair. That is, I deliberately constructed a family of integers that were known to be a-priori rough. But, suppose I gave you some large, rather arbitrarily constructed number, and asked you to tell me if it is prime? For example, to pull a number out of my hat, consider
P = sym(2)^122397 + 65;
floor(vpa(log10(P) + 1))
36846 decimal digits is pretty large. And in fact, large enough that sym/isprime in R2024b will literally choke on it. But is it prime? Can we efficiently learn if it is at least not prime?
A nice way to learn the roughness of even a very large number like this is to use GCD.
gcd(P,prod(sym(primes(10000))))
If the greatest common divisor between P and prod(sym(primes(10000))) is 1, then P is NOT divisible by any small prime from that set, since they have no common divisors. And so we can learn that P is indeed fairly rough, 10000-rough in fact. That means P is more likely to be prime than most other large integers in that domain.
gcd(P,prod(sym(primes(100000))))
However, this rather efficiently tells us that in fact, P is not prime, as it has a common factor with some integer greater than 1, and less then 1e5.
I suppose you might think this is nothing different from doing trial divides, or using the mod function. But GCD is a much faster way to solve the problem. As a test, I timed the two.
timeit(@() gcd(P,prod(sym(primes(100000)))))
timeit(@() any(mod(P,primes(100000)) == 0))
Even worse, in the first test, much if not most of that time is spent in merely computing the product of those primes.
pprod = prod(sym(primes(100000)));
timeit(@() gcd(P,pprod))
So even though pprod is itself a huge number, with over 43000 decimal digits, we can use it quite efficiently, especially if you precompute that product if you will do this often.
How might I use roughness, if my goal was to find the next larger prime beyond 2^122397? I'll look fairly deeply, looking only for 1e7-rough numbers, because these numbers are pretty seriously large. Any direct test for primality will take some serious time to perform.
pprod = prod(sym(primes(10000000)));
find(1 == gcd(sym(2)^122397 + (1:2:199),pprod))*2 - 1
2^122397 plus any one of those numbers is known to be 1e7-rough, and therefore very possibly prime. A direct test at this point would surely take hours and I don't want to wait that long. So I'll back off just a little to identify the next prime that follows 2^10000. Even that will take some CPU time.
What is the next prime that follows 2^10000? In this case, the number has a little over 3000 decimal digits. But, even with pprod set at the product of primes less than 1e7, only a few seconds were needed to identify many numbers that are 1e7-rough.
P10000 = sym(2)^10000;
k = find(1 == gcd(P10000 + (1:2:1999),pprod))*2 - 1
k =
Columns 1 through 8
15 51 63 85 165 171 177 183
Columns 9 through 16
253 267 273 295 315 421 427 451
Columns 17 through 24
511 531 567 601 603 675 687 717
Columns 25 through 32
723 735 763 771 783 793 795 823
Columns 33 through 40
837 853 865 885 925 955 997 1005
Columns 41 through 48
1017 1023 1045 1051 1071 1075 1095 1107
Columns 49 through 56
1261 1285 1287 1305 1371 1387 1417 1497
Columns 57 through 64
1507 1581 1591 1593 1681 1683 1705 1771
Columns 65 through 69
1773 1831 1837 1911 1917
Among the 1000 odd numbers immediately following 2^10000, there are exactly 69 that are 1e7-rough. Every other odd number in that sequence is now known to be composite, and even though we don't know the full factorization of those 931 composite numbers, we don't care in the context as they are not prime. I would next apply a stronger test for primality to only those few candidates which are known to be rough. Eventually after an extensive search, we would learn the next prime succeeding 2^10000 is 2^10000+13425.
In my next post, I show how to use MOD, and all the cores in your CPU to test for roughness.

