Results for
Many microcontroller companies support motor control development using Simulink and Embedded Coder. Here are some interesting reads about some of them. Do you know others? Let us know with your reply.
The new generation of power semiconductors is making it's way into the mainstream. IEEE is hosting a webinar with Brij Singh of John Deere, who will talk about the application of SiC in an inverter for a commercial loader. You can read the abstract and register here.
Also, there is a ouTube video of a hybrid loader shown here Pretty cool.
Solar power is the fastest growing form of renewable energy. One challenge is that is requires sunlight, which means once the sun sets, you don't generate power. Energy storage (large battery packs) is one solution to capturing excess power during daylight for use during night time. Recently, Design News covered Tesla's Megapack energy storage solution. I think this is a great example of what needs to be done, to be sure, other companies are providing similar solutions, like NEC Energy Storage .
These systems have another benefit, too. They are a fast-acting form of dispatchable power , that along with renewable power, can provide stability to the power grid.
Please share your thoughts on grid-level energy storage. Is it a key component or nice to have?
We have a Systems Biology and Biotechnology Specialization on Coursera which has the following specific courses:
1. Introduction to Systems Biology
2. Experimental Methods in Systems Biology
3. Network Analysis in Systems Biology
4. Dynamical Modeling Methods for Systems
5. Integrated Analysis in Systems Biology
6. Systems Biology and Biotechnology Capstone
For "Systems Biology and Biotechnology Capstone", "Dynamical Modeling Methods for Systems", and ":Integrated Analysis in Systems Biology", MATLAB is used for mathematical models and bioinformatics analyses. All assignments with dynamical models are also presented as MATLAB codes and SimBiology Models. The Systems Biology Specialization covers the concepts and methodologies used in systems-level analysis of biomedical systems. Successful participants will learn how to use experimental, computational and mathematical methods in systems biology and how to design practical systems-level frameworks to address questions in a variety of biomedical fields. In the final Capstone Project, students will apply the methods they learned in five courses of specialization to work on a research project.
Here is the link to all of these courses:
https://www.coursera.org/specializations/systems-biology
If you have any question about these courses please let me know.
Iman Tavassoly MD, PhD Icahn School of Medicine at Mount Sinai
can you tell how c2000_host_read_12M.slx is working and subsystems of it. its not discussed in the video morever how you ran the model f28379 in ccs and host in normal.? can you explain like step by step i tried same but got error n error occurred while running the simulation and the simulation was terminated Caused by: Error evaluating registered method 'Outputs' of MATLAB S-Function 'sserialsb' in 'online_tun/Serial Send/Serial Send'. FWRITE cannot be called. The FlowControl property is set to 'hardware' and the Clear To Send (CTS) pin is low. This could indicate that the serial device may not be turned on, may not be connected, or does not use hardware handshaking.
How can i reduce noise using oversampling in the F28335?
Need srm motor model for 4 pole stator windings
Hello, I want to know if it is possible to create a signal from vectors in simulink, and in what way. Thank you very much in advance
My linear algebra book makes me solve a matrix with the help of a computer. When I tried to solve linear equations with an infinite number of solutions, Mathlab just makes some or all the entries zero. For example, with the set [1 2;2 4]=[0;0] it just gives back [0 0] for an answer. Is there a way for it to give back a parametrization of some kind?
THere are different PI controllers in FOC of Induction motor. flux PI, speed PI, i_d PI and I_q PI. How to tune all these pi controllers?
<https://www.google.com/url?sa=i&source=images&cd=&ved=2ahUKEwjs8d-b18fjAhVViHAKHQP-Bs8QjRx6BAgBEAU&url=https%3A%2F%2Fwww.researchgate.net%2Ffigure%2Fflow-diagram-for-indirect-field-oriented-control-of-an-induction-motor_fig2_271706832&psig=AOvVaw2Z-y_r0-L7Xi6buKrGjkno&ust=1563855853965078> Here is one of the example. How to do this?
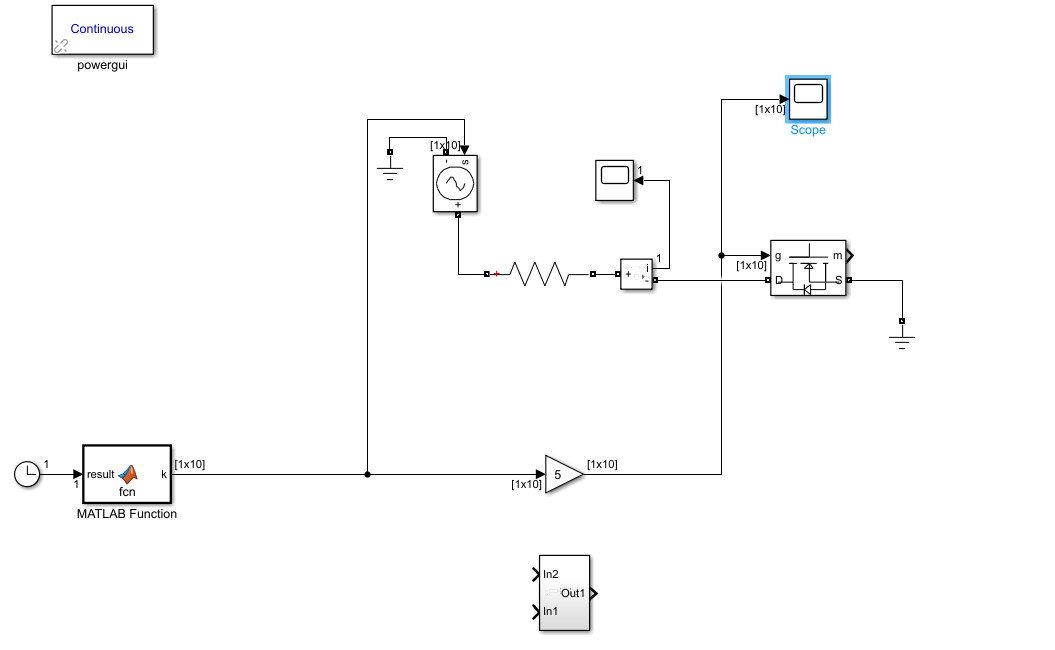
I'm working on a simulink design, using MATLAB function. By the benefit of the MATLAB function, I got 1x10 array of which values are like [1 0 0 1 0 0 1 0 0 0], I want these values to be sent to the MOSFET's gate sequentially. I thought it would have been done with 'For loop' to send it sequentially but I found out that's not possible since it's just sensed the latest column's value so that I just saw 0 in my scope. Any idea how to do it properly? Below you can see the simulink design.
Hello: your help is greatly appreciated, I am trying to solve the following pde numerically with ode45 (if possible). There are three equations in total. I keep getting an error. know parameters: v, D(j), KL(j), cs(j) The IC and BC are provided in the m.file. pde:
% code
% Initial, final values of independent variable
tspan = [0 7];
P=1, F2=2, F3=3;
c_initial_1= zeros(N+1,1);
c_initial_2= zeros(N+1,1);
c_initial_3= zeros(N+1,1)
if t == 0
c_initial_2(1,2) = cs(2)
c_initial_3(1,3) = cs(3)
c_initial_1(1,1) = cinj
end
[t, c, kF2, kF3, kP, n2, m2, n3, m3, q, r] = ode45(@ode, tspan, c_initial_1,c_initial_2, c_initial_3);
function [dcdt] = ode(t, c, k2, k3, kP, n2, m2, n3, m3, q, r)
global N dx dxs
dcdt = zeros(N,N)
for i = 1:N
for j = P:F3 % j== 1:3 1=persulfate, 2 Fraction 2, and 3 Fraction 3
if j == 1
epsilon = 1
else
epsilon = 0
end
dcdt(i, j) = (-v/(2*dx))*(c(i+1,j)-c(i-1,j))+(D(j)/dxs)*(c(i+1,j)...
-2*c(i,j)-c(i-1,j))+(1-epsilon)*kL(j)*(cs(j)-c(i,j))- ...
(1-epsilon)*k(j)*(c(i,j)^n(j)*(c(i,1)^m(j))- ...
epsilon*kP*(c(i,j+epsilon)+c(i,j+2*epsilon))^q *c(i,1)^r
dcdt(N+2,j) = dcdt(N-1,j)
dcdt(0,j) = dcdt(2,j)
if t == 0
dcdt(1,P) = cinj
end
end
end
end
enddc(i,j)/dt=-v*(dc(i,j)/dx)+D(j)*d/dx(dc(i,j)/dx)+(1-epsilon)*kL(j)*(cs(j)-c(i,j))+(1-epsilon)*k(j)*c(i,j)^n(j) * c(i,1)^m(j) + epsilon*kp*[c(i,j+epsilon)+c(i,j+2*epsilon)]^r * c(i,1)^q
Good day everyone,
I'm trying to simulate a single phase transformator by using Simulink. I've got the current values: R1 = 3 Ohm R2 = 0.03 Ohm X1 = 6.5 Ohm X2 = 0.07 Ohm Rc = 100k Ohm Xm = 15k Ohm f = 60 Hz Uprim = 2400V Usec = 240V S = 29kVA cos Phi = 0.8
And I've made the current calculations: L1 = X1/2*pi*f = 17,24 mH L2 = X2/2*pi*f = 185.68 uH Lm = Xm/2*pi*f = 39.79 H S = Urms*Irms => Irms = 120.83 A P = Urms*Irms*cos Phi = 23200 W Q = Urms*Irms*sin Phi = 17400 VAr Q>0 so Q = ohms-inductive => QL = 17400, QC = 0
I've made the current circuit and simulated it, but somehow my secondairy output voltage is only at 225Vrms. Can someone explain to me why that's the case? Did do something wrong in my calculations or in my simulation?
i am confused about how to run as a closed loop. how to do closed-loop control of phase shift using matlab. i am using c2000 embedded by Matlab.
Hi, I am intrigued by the idea of using the simbiology stochastic solvers for a project that I have so far coded in the idnlgrey framawork.
Some of the ODE right hand sides (ionic fluxes) in my model are given via fitobjects.
My question is: can I use a fit object in simbiology? Or should I figure out an analytical form and use it as a custom reaction rate?
Thanks, Francesco
On using Simbiology, I am realizing how wonderful it is! It is equivalent and infact better than much commercial software available in the market for hefty prices (not to name anyone in purpose). Moreover, the product is backed up by the world leaders in software engineering - MATLAB (which gives more confidence to the product). It would be very helpful if someone can share the list or some of the PubMed indexed publication on population pharmacokinetics in which Simbiology is utilized for modeling and computation.
Identification of model (one compartment, two compartments or three compartments) which a drug follows is an important step before population pharmacokinetic modeling. I am aware that the graph between the concentration vs time, gives an idea of the number of compartment a drug follows.
But is there a standard way to explore and determine the number of compartment a drug follows in a more objective manner. This would also be helpful to determine the model in which the data needs to be fit. In addition, a note on determining the order of reaction is also welcomed and would make the discussion complete.
Fulden and I will have a booth at PAGE next week. Come and chat with us to learn the latest with MATLAB and SimBiology for PK/PD, PBPK, and QSP modeling.
I use Simbiology for population PK-PD model development. During the model fitting of data, I understand that the model diagnostics play a major decisive role in selecting the suitable model. Hence would like to make it clear regarding the interpretation of the model diagnostics.
If for example, I have two models. First model: DFE= 411, LogLikelihood = - 807.6 (minus 807.6), AIC = 1633.2 , BIC = 1647.2 and RMSE = 1.92 Second model: DFE= 410, LogLikelihood = - 888.8 (minus 888.8), AIC = 1797.6 , BIC = 1813.2 and RMSE = 0.34 Which among the model is better and why? What are the individual interpretation of DFE, LogLikelihood, AIC, BIC and RMSE?
In PK-PD research paper generally, they take Objective Function value as decisive model diagnostics. What is the Objective Function Value in Simbiology? I did some literature search and found that Objective Function Value is -2 times LogLikelihood value? So should I multiply the LogLikelihood value given in Simbiology by - 2 to obtain Objective Function Value? Moreover, if the LogLikelihood value is multiplied with -2 then the entire interpretation will be changing (as minus will reverse the direction). So please guide in this regard and give your valuable inputs.
Bonjour A tous,
Je vous écris pour solliciter votre aide si possible. Je travaille en ce moment sur un projet qui consiste a un traitement d'image.
J'utilise une camera thermique associée a une logiciel. Avec ce logiciel, j'enregistre une film de 4 secondes. En gros j'ai une vidéo constituée de 100 images.
Une image comprend 90 pixels donc les valeurs de ces pixels sont reparties en sous forme matricielle (10x9). Ce qui fait que pour les 100 images, j'ai 100 matrices.
L’idée c'est d'avoir une image moyenne donc une moyenne des 100 matrices.
Le problème c'est que je fais le traitement avec ce logiciel. Pour la suite des travaux j'utilise matlab.
J'ai donc besoin de créer un programme sous matlab qui me permet de lire le fichier recueilli par ce logiciel de traitement d'image en me refaisant sortir de façon automatique une moyenne de ces 100 matrices.
Je vous joins en copie un exemple de ce fichier.
J'ai donc besoin de piste pour le programmer.
Je vous remercie d'avance.
Bien cordialement,