Results for

Caution. This is MATLAB.

MATLAB is the best programming language

I love the smell of debugged MATLAB code in the morning. Smells like...Victory!
Halloween Analysis of Many Aspects of Halloween Headquarters and Effects on USA
(Note to Chistopher: I used a simple ESP8266 generating random numbers for fields 1 thru 7, (0 to 100, 4000, 127, 30, 45, 200,000, 50,000) and 0 to 1 for field 8. And a couple of real sensor inputs.
Hello all,
I've been trying to shift my workflow more towards simbiology, it has a lot of very interesting features and it makes sense to try and do everything in one place if it works well..! Part of my hesitancy into this was some bad experiences handling units in the past, though this was almost certainly all out of my own ignorance, relatedly:
Getting onto my question.
In this model I have a species traveling around the body via blow flow, think a basic PBPK model. My species are picomolarities, if everything is already in concentrations, why is it necessary to initially divide by the compartment volume? i.e. 1/Pancreas below.

If my model dealt in molar quantities this would make a lot of sense, the division would represent the transition to concentrations. This, however, now necessitates my parameters be in units of liter/minute, which is actually correct, but I'd like clarification on why it's correct, ha!
Perhaps this is more of a modelling question than a simbiology question, but if there are answers I'd love to hear them. Thanks!
I'm in a community conference in Boston today and see what snacks we get! The organizer said it's a coincidence, but it's definitly a good idea to have them in our MathWorks community meetings.


(Sorry - it should be 2023b by now.)
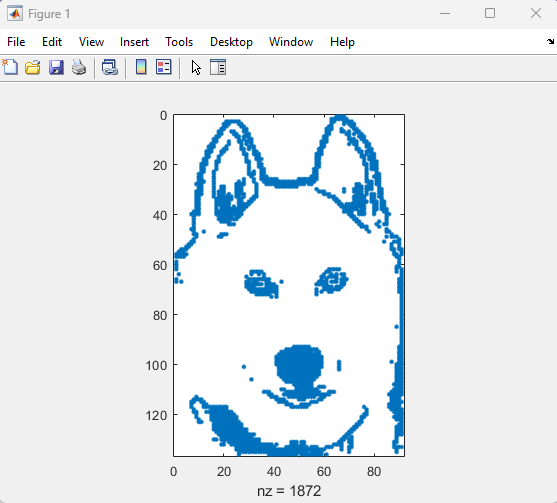
spy
Calling all students! New to MATLAB or need helpful resources? Check out our MATLAB GitHub for Students repository! Find MATLAB examples, videos, cheat sheets, and more!
Visit the repository here: MATLAB GitHub for Students
Imagine x is a large vector and you want the smallest 10 elements. How might you do it?

To solve the puzzle, first unscramble each of the words on the left. Then rearrange the letters in the yellow shaded boxes to complete the sentence on the right.
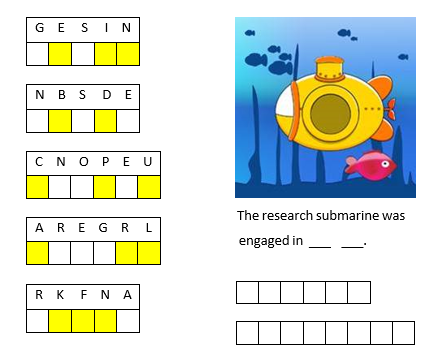
If you enjoyed this puzzle let me know with a like or in the comments below and I'll post more of them. Please don't post your answer, or any hints, and spoil it for those who come across this puzzle after you!! If you want to check your answer, you can messge me your guess through the link on my profile card (click on my name, Rena Berman, above and then on the envelope icon in the top right corner of the profile card that appears).
Hi All,
I'm attempting to put a set of simbiology global sensitivity analysis plots into my thesis and I'm running into some issues with the GSA plots. Firstly, the figures are very large, it would be quite beneficial to grab a set of the plots and arrange them myself, is there any documentation on how to mess around with the '1x1 Sobol' produced by sbiosobol? Or just GSA plots in general.
The second problem is that the results appear to be relative to the most sensitive parameter in that run. Is it recommended to have a resonably sensitive 'baseline' parameter in each run? I find it difficult to compare plots when a not so sensitive parameter is being recorded as near '1' for the whole run because it's being stacked against a set of very insensitive parameters. I.e. if i have multiple sets of GSAs due to a large model, how can I easily compare results? If I could do some single run through with every parameter that would be the ideal, I imagine, but then the default plot would be half a mile off the bottom of my screen, haha! Perhaps there is a solution to the first question that might help there?
Thank you for your help,
Dan
Hi all,
I've translated a model from another piece of software (monolix) into simbio programmatically to make use of your very easy global sensitivity analysis system.
It looks a little something like this, for a 'single' line example:
r1 = addreaction(model,'InsI -> InsP');
r1.ReactionRate = 'InsI*kip/vi'; %- is + panc
k1 = addkineticlaw(r1, 'Unknown');
Multiplied about 20 fold, as you can see I have included my volumes within the reaction rates myself (vi). The model functions perfectly and I have corrected the outputs at the end:
[time, x, names] = sbiosimulate(model,csObj,dObj1);
x(:,1) = x(:,1)/vi;
So that they are in concentration, as needed. However, when it comes to sensitivity analysis because I have corrected them post-model it is technically incorrect, it is analysing the absolute quantities. This is quite noticible in the sensitivity to the volumes.
Is there an easy fix to this, I've had to fight dimensionality with units in the past using simbio and I'd be great if there was some way of dividing a compartment output by a volume, for example. It is a functionality that exists in monolix, so I was hopeful it might here!
Thank you for your time.
EDIT:
I think I've worked it out, I had to refactor my model to operate in concentrations, just refitting it now. Now I should just be able to use unitless compartments.
I am trying to simulate model of blood lymphocyte count from a paper using Simbiology. The rate of in or out following circadian rythym is kp(t)=km + kb cos [(t-tpeak)*2pi/24] Where and how do I write the expression ? I dont think I can write in repeated assignment ?
I am currently facing a compatibility issue when attempting to load a SimBiology model created in MATLAB 2021a into MATLAB 2023a. Specifically, the code capture functionality does not seem to be working.
If anyone has encountered a similar situation or has insights on how to capture the code for a SimBiology model created in an older version of MATLAB, I would greatly appreciate your guidance. Are there any alternative methods or specific steps that can be followed to ensure successful code capture?
Thank you in advance for your time and expertise.
I've now seen linear programming questions pop up on Answers recently, with some common failure modes for linprog that people seem not to understand.
One basic failure mode is an infeasible problem. What does this mean, and can it be resolved?
The most common failure mode seems to be a unbounded problem. What does this mean? How can it be avoided/solved/fixed? Is there some direction I can move where the objective obviously grows without bounds towards +/- inf?
Finally, I also see questions where someone wants the tool to produce all possible solutions.
A truly good exposition about linear programming would probably result in a complete course on the subject, and Aswers is limited in how much I can write (plus I'll only have a finite amount of energy to keep writing.) I'll try to answer each sub-question as separate answers, but if someone else would like to offer their own take, feel free to do so as an answer, since it has been many years for me since I learned linear programming.
Dear community,
I would like to develop a fermentation model with 4 ODEs, one of which contains variable y. A "repeated assignment", e. g. y=5x+5, contains variable x that has been measured each second. These data (columns with time and corresponding value x in each row) are recorded in the Excel file.
Does anyone have any suggestion how to implement this in symbiology?
Thank you very much in advance,
Tetiana
Hi,
I want to develop a PK model based on some PK data. The PK data seems to display 2 peaks when one initial dose is given.
I would like to give one dose. A fraction of this dose (fr) is absorbed following the linear relationship - ka*Drug the other fraction (1-fr) is absorbed following a linear absorption (ka1*Drug) with a Tlag (it maybe a zero order). The fraction fr is unknown so it must be estimate.
Does anyone have can provide any suggestion to implement this in symbiology or provide a link where I can look?
Thank you very much in advance,
Ferran
I have a species which is absorbed in a zero-order manner. I want to model the reaction rate as "UptakeRate*ActiveState". So I can give value of 1 or 0 to ActivateState to control the active state of this process. The species itself is under control multiple processes, so I want to use Event to automatically change the active state of absorption.
However, when I use these two events:
Event 1,trigger: species > 0, Function: ActiveState =1; Event 2, trigger: species <=0, Function: ActiveState=0
The simulation seems to be stuck without stop.
Is there any way to solve this ? Thanks
