Apply Transfer Learning on PyTorch Model to Identify 5G and LTE Signals
This example shows how to use a pretrained PyTorch® semantic segmentation neural network to identify 5G NR and LTE signals in MATLAB®. MATLAB generates and preprocesses standard-based wireless signals for training and testing.
Introduction
Computer vision uses the semantic segmentation technique to identify objects and their locations in an image or a video. In wireless signal processing, the objects of interest are wireless signals, and the locations of the objects are the frequency and time occupied by the signals. In this example, you apply the semantic segmentation technique to wireless signals to identify spectral content in a wideband spectrogram.
In this example, you:
Set up your Python® environment to call PyTorch functions from MATLAB.
Load a pretrained PyTorch network into MATLAB.
Generate, augment, and preprocess 5G NR and LTE signals online to fine-tune the PyTorch model weights.
Evaluate and visualize the network outputs in MATLAB.
Associated AI for Wireless Examples
You can use this example as part of a complete deep learning workflow:
Capture and label bandwidths
The Capture and Label NR and LTE Signals for AI Training (Wireless Testbench) example shows how to scan, capture, and label bandwidths with 5G NR and LTE signals using an SDR.
Train Network with labeled bandwidths
The following examples show how to train a semantic segmentation network to identify 5G NR and LTE signals in a wideband spectrogram.
The Spectrum Sensing with Deep Learning to Identify 5G, LTE, and WLAN Signals shows how to train a semantic segmentation network in MATLAB from scratch or apply transfer learning to a pretrained network.
This example shows how to use the Python interface in MATLAB to fine-tune a pretrained PyTorch™ semantic segmentation network using transfer learning.
Identify 5G and LTE signals using SDR
The Identify LTE and NR Signals from Captured Data Using SDR and Deep Learning (Wireless Testbench) example shows how to use a deep learning trained semantic segmentation network to identify NR and LTE signals from wireless data captured with an SDR.
For more information on implementing AI workflows for wireless communications, see AI for Wireless.
Set Up Python Environment
Before running this example, set up the environment as explained in PyTorch Coexecution. Specify the full path of the Python executable to use in the executablePath field. The helperSetupPyenv function sets the Python environment in MATLAB according to the selected executionMode and checks that the libraries listed in the requirements_specsense.txt file are installed.
executionMode ="OutOfProcess"; reqFileName = "requirements_specsense.txt"; if ispc executablePath =
".venv\Scripts\pythonw.exe"; else executablePath =
"venv/bin/python"; end currentPyenv = helperSetupPyenv(executablePath,executionMode,reqFileName);
Setting up Python environment Parsing requirements_specsense.txt Checking required package 'torch' Checking required package 'torchvision' Required Python libraries are installed.
Deeplabv3 Semantic Segmentation Network
This example uses the pretrained deeplabv3 semantic segmentation network. The example modifies the deeplabv3 to set the number of output classes to three: "Noise", "NR", and "LTE". Set the network backbone as resnet50 or resnet101.
classNames = ["Noise" "NR" "LTE"]; networkBackbone ="resnet50";
The deeplabv3nn.py module is a helper Python module that contains all the functions required to train and test the PyTorch neural network for this example. The PyTorch Wrapper Template sections shows how to create the interface functions using a template. Display short information on the model using the info function.
numClasses = length(classNames);
model_size = py.deeplabv3nnwrapper.info(networkBackbone,numClasses);
numLayers = single(model_size{1});
numLearnables = single(model_size{2})/1e6;
disp("Deeplabv3 model with "+networkBackbone+...
" backbone has "+num2str(numLayers)+" layers and "+...
num2str(numLearnables)+" M learnables.");Deeplabv3 model with resnet50 backbone has 189 layers and 41.9948 M learnables.
Specify Data Set Parameters
This example follows the steps in Generate Training Data section in the Spectrum Sensing with Deep Learning to Identify 5G, LTE, and WLAN Signals example to generate a data set of 5G NR and LTE frames. Each sample of the data set contains a frame containing either a 5G NR signal, an LTE signal, or a combined 5G NR and LTE signal using randomly selected values from the standards parameter set specified below. Each frame passes through a fading channel model with randomly selected SNR and Doppler shift.
params.Fs = 61.44e6; % Hz params.NumSubFrames = 40; % corresponds to 40 ms % 5G NR Parameters params.SCSVec = [15 30]; params.BandwidthVec = [10:5:30 40 50]; % [5:5:25 30:10:100] params.MaxTimeShift = params.NumSubFrames; % Time shift in milliseconds params.SSBPeriod = 20; % [5 10 20 40 80 160] 20 is most frequently found OTA % LTE Parameters params.RCVec = ["R.2","R.6","R.8","R.9"]; params.TrBlkOffVec = [1,2,3,4,5,6,7,8]; % Channel Parameters params.SNRMin = 0; % dB params.SNRMax = 40; % dB params.DopplerMin = 0; % Hz params.DopplerMax = 500; % Hz params.CarrierFreq = 4e9; % Hz
To train a semantic segmentation model for spectrum sensing, generate the power spectral density (PSD) spectrogram of the impaired signal and convert it into a three-channel image. Each pixel in the spectrogram image belongs to one of three classes, that is, Noise, NR, or LTE. An advantageous loss function for handling pixel class imbalance when training semantic segmentation networks is the Dice loss function.
The BCE-Dice loss between the network output, Y, and a pixel label target, T, is defined as
where
The helperSpecSenseGenerateImagesMasks function generates a spectrogram image and the corresponding pixel mask as a binary 3-D array, where the first two dimensions are equal to the height and width of the the spectrogram image and the third dimension is the number of classes in the data set. Specify the spectrogram image parameters.
% Spectrogram Parameters params.NOverlap = 10; params.Window = hann(256); params.Nfft = 4069; % Image and mask parameters params.ImageSize = [256 256]; % pixels params.ClassNames = classNames; params.NumStandards = 2;
Train Deep Neural Network
In this section, you train the PyTorch Deeplabv3 model using the Python interface with run-time data generation and preprocessing in the training loop. During each training iteration, you:
Generate a set of 5G and LTE signals.
Augment each 5G/LTE signal with multiple fading channel realizations to create a
miniBatchof training samples.Preprocess the augmented time domain signals.
Train the model for one iteration.
Data augmentation is a technique that allows you to increase the size of a training data set by adding random impairments without generating or collecting new data signals. Augmenting the training data often improves the network generalization. In this example, the impairmentsPerSignal parameter specifies the ratio between the number of frames generated from standard to the number of channel-impaired signals. The default value of impairmentsPerSignal is set to 2, that is, each 5G NR/LTE frame is impaired with two randomly generated fading channel realizations. This figure shows the augmented data samples resulting from one 5G NR signal and one LTE signal.
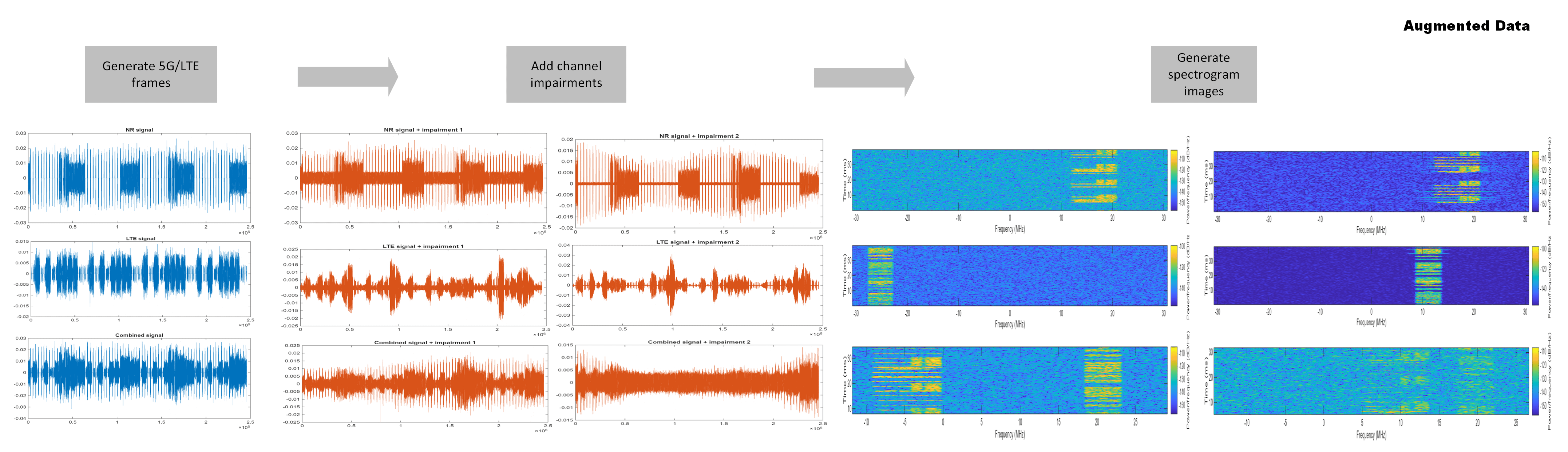
impairmentsPerSignal =  2;
miniBatch = 30;
2;
miniBatch = 30;Set trainNow to true to train the model. Training the model for 500 iterations with background data generation takes approximately 1 hour and 20 minutes using an Intel® Xeon® W-2133 CPU and an NVIDIA® RTX 3080 GPU. If trainNow is set to false, the example loads a pretrained model to run the example quickly.
trainNow =false; plotLoss =
false; verbose =
true; valFreq = 10; saveCheckPointFreq = 50; maxTrainIter = 500; params.MiniBatchSize = miniBatch; params.ImpPerSig = impairmentsPerSignal;
Train Network with Background Data Generation
To train the PyTorch model with run-time data generation, in each training iteration, you generate a minibatch of augmented time-domain frames using helperSpecSenseGenerateSignals function. Then, you generate the spectrogram and masks of each time domain signal in the minibatch using helperSpecSenseGenerateImagesMasks function. Use the deeplabv3_object to compute the loss function value and update the PyTorch model weights.
if trainNow weightsFileName = "trained_specsenselabv3.pth"; %#ok deeplabv3Obj = py.deeplabv3nnwrapper.setup_trainer( ... networkBackbone, ... weightsFileName, ... saveCheckPointFreq, ... maxTrainIter);
backgroundDataGen is set to true by default to speed up the training. You use Parallel Computing Toolbox™ to generate and augment each minibatch of time domain signals using a background parallel pool. If the Parallel Computing Toolbox is not available, this section runs on a single CPU worker.
Use the helperGetDataBatch function to generate a batch of augmented signals using a background parallel pool. Use the CPU or the GPU, if one is available, to generate the spectrogram images and masks for the batch of augmented signals and train the model for one iteration.

backgroundDataGen =true; tstart = tic; if backgroundDataGen & canUseParallelPool [trainFuture,valFuture] = helperSetBackgroundDataGen(params,"train"); for iteration = 1:maxTrainIter [trainFuture,images,masks] = helperGetDataBatch(params,trainFuture); py.deeplabv3nnwrapper.train_one_iteration(deeplabv3Obj,images,masks); if mod(iteration,valFreq)==0 || iteration == 1 [valFuture,images,masks] = helperGetDataBatch(params,valFuture); py.deeplabv3nnwrapper.validate(deeplabv3Obj,images,masks); t = seconds(toc(tstart)); t.Format = "hh:mm:ss"; helperTrainingProgress(iteration,maxTrainIter,deeplabv3Obj, ... verbose,plotLoss,valFreq,t); end end else
Train Network with In-Process Data Generation
Set backgroundDataGen to false to generate the data, preprocess the data, and train the network using the same process. Generating the data in-process increases the training time to approximately 4 hours and 30 minutes using an Intel Xeon W-2133 CPU and an NVIDIA RTX 3080 GPU.
for iteration = 1:maxTrainIter [signals,freqLim,labels] = helperSpecSenseGenerateSignals(params); [images,masks] = helperSpecSenseGenerateImagesMasks(params,signals,freqLim,labels); py.deeplabv3nnwrapper.train_one_iteration(deeplabv3Obj,images,masks); if mod(iteration,valFreq)==0 [signals,freqLim,labels] = helperSpecSenseGenerateSignals(params); [images,masks] = helperSpecSenseGenerateImagesMasks(params,signals,freqLim,labels); py.deeplabv3nnwrapper.validate(deeplabv3Obj,images, masks); t = seconds(toc(tstart)); t.Format = "hh:mm:ss"; helperTrainingProgress(iteration,maxTrainIter,deeplabv3Obj, ... verbose,plotLoss,valFreq,t); end end end else weightsFileName = "specsenselabv3.pth"; helperDownloadFiles(); deeplabv3Obj = py.deeplabv3nnwrapper.construct_model( ... networkBackbone, ... weightsFileName); end
Starting download of data files from: https://www.mathworks.com/supportfiles/spc/SpectrumSenseTorchModel/SpectrumSenseLabV3.zip Extracting files. Extract complete.
Test Deep Neural Network
Test the trained PyTorch model using a new set of generated data with 300 impaired 5G NR and LTE signals. The example specifies the size of the test data set using the product of the minibatch and numTestIter variables.
numTestIter = 10; backgroundDataGen = true; numTestSamples = miniBatch*numTestIter
numTestSamples = 300
disp("Testing model ...")Testing model ...
if backgroundDataGen testFuture = helperSetBackgroundDataGen(params,"test"); for iteration = 1:numTestIter [testFuture,images,masks] = helperGetDataBatch(params,testFuture); py.deeplabv3nnwrapper.test(deeplabv3Obj,images,masks); if mod(iteration,2)==0 disp("Tested "+num2str(iteration*miniBatch)+" signals ...") end end else for iteration = 1:numTestIter %#ok [signals,freqLim,labels] = helperSpecSenseGenerateSignals(params); [images,masks] = helperSpecSenseGenerateImagesMasks(params,signals,freqLim,labels); py.deeplabv3nnwrapper.test(deeplabv3Obj,images,masks); if mod(iteration,5)==0 disp("Tested "+num2str(iteration*miniBatch)+" signals ...") end end end
Tested 60 signals ... Tested 120 signals ... Tested 180 signals ... Tested 240 signals ... Tested 300 signals ...
disp("Testing model with "+num2str(numTestSamples)+" images complete.")
Testing model with 300 images complete.
IOU = deeplabv3Obj.test_iou;
Accuracy = deeplabv3Obj.test_accuracy;
numTrainSamples = miniBatch*maxTrainIter;
resultsTable = table(categorical({'Deeplabv3_resnet50'}), ...
numTrainSamples,numTestSamples,IOU,Accuracy, ...
VariableNames=["Network" ...
"Training Dataset Size" ...
"Test Dataset Size" ...
"Mean IOU" ...
"Mean Accuracy"]);
disp(resultsTable) Network Training Dataset Size Test Dataset Size Mean IOU Mean Accuracy
__________________ _____________________ _________________ ________ _____________
Deeplabv3_resnet50 15000 300 0.80555 99.472
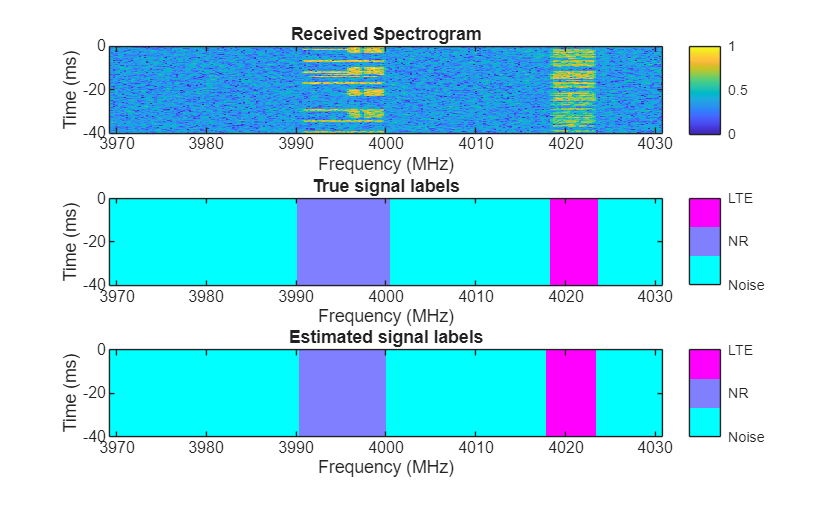
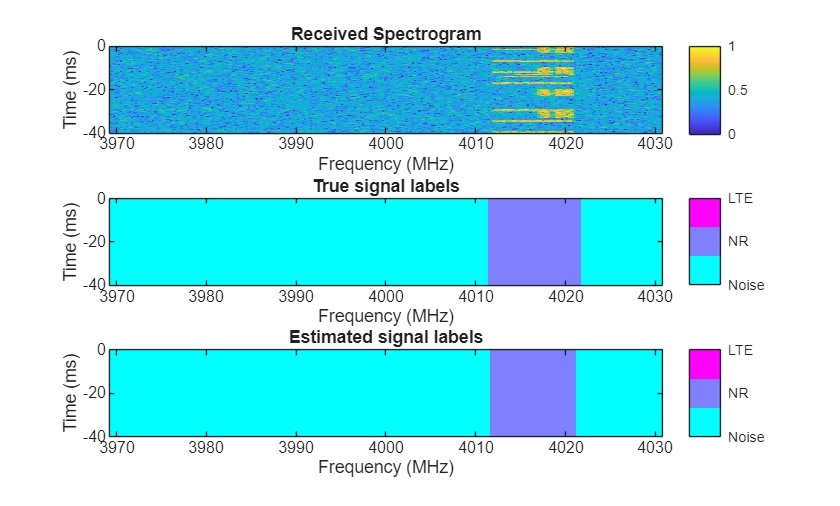
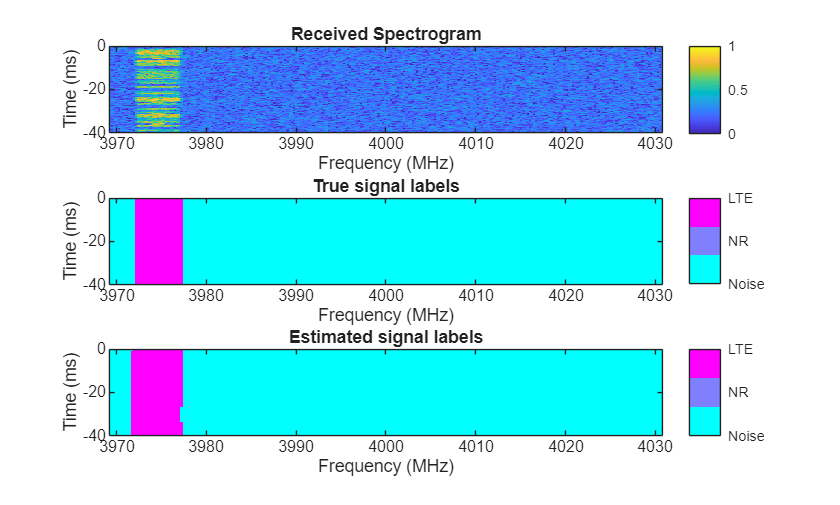
Visually examine the classification accuracy of the trained PyTorch network using randomly generated test data samples.
rng(10) frameDuration = params.NumSubFrames*1e-3; % seconds params.MiniBatchSize = 3; params.ImpPerSig = 1; [signals,freqLim,labels] = helperSpecSenseGenerateSignals(params); [images,masks] = helperSpecSenseGenerateImagesMasks(params,signals,freqLim,labels); predictions = single(py.deeplabv3nnwrapper.predict(deeplabv3Obj,images)); for idx = 1:3 rcvdSpectrogram = squeeze(images(:,:,:,idx)); trueLabels = categorical(ones(256),1:length(params.ClassNames),params.ClassNames); predictedLabels = categorical(ones(256),1:length(params.ClassNames),params.ClassNames); for i = 2:3 trueLabels(squeeze(masks(:,:,i,idx))==1) = categorical(params.ClassNames(i)); predictedLabels(squeeze(predictions(:,:,i,idx))==1) = categorical(params.ClassNames(i)); end figure helperSpecSenseDisplayResults(im2uint8(rescale(rcvdSpectrogram)),trueLabels,predictedLabels, ... params.ClassNames,params.Fs,params.CarrierFreq,frameDuration); end
Helper Files
helperdeeplabv3nn.py
helperdeeplabv3nnwrapper.py
helperinstalledlibs.py
helperLibraryChecker.m
helperSetupPyenv.m
helperSpecSenseDisplayResults.m
helperSpecSenseGenerateImagesMasks.m
helperSpecSenseGenerateSignals.m
helperSpecSenseLTESignal.m
helperSpecSenseNRSignal.m
PyTorch Wrapper Template
You can use your own PyTorch models in MATLAB using the Python interface. The py_wrapper_template.py file provides a simple interface with a predefined API. This example uses the following API set:
construct_model: returns the PyTorch neural network modelsetup_trainer: sets up a trainer object for with online trainingtrain_one_iteration: trains the PyTorch model for one iteration for online trainingvalidate: validates the PyTorch model for online trainingpredict: runs the PyTorch model with the provided input(s)test: Performs a test step with the given images and masks.
info: prints or returns information on the PyTorch modelprocess_data_from_matlab: Processes image and mask data from MATLAB format to torch tensors.process_data_to_matlab: Processes network output from torch tensors to a format suitable for post processing in MATLAB.
The Train PyTorch Channel Prediction Models example shows an offline training workflow and uses the following API set in addition to the ones used in this example.
train: trains the PyTorch modelsave_model_weights: saves the PyTorch model weightsload_model_weights: loads the PyTorch model weights
You can modify the py_wrapper_template.py file. Follow the instruction in the template file to implement the recommended entry points. Delete the entry points that are not relevant to your project. Use the entry point functions as shown in this example to use your own PyTorch models in MATLAB.
Local Functions
function varargout = helperSetBackgroundDataGen(params,status) %helperSetBackgroundDataGen Sets a background pool and % starts the data generation in the background. if ~isempty(gcp("nocreate")) p = gcp("nocreate"); else currentCluster = parcluster; p = parpool(min(currentCluster.NumWorkers,8)); end if strcmp(status,"train") params.numWorkersTrain = floor(p.NumWorkers*0.8); numTrainFutures = max(params.numWorkersTrain*3,12); varargout{1}(1:numTrainFutures) = parallel.FevalFuture; for idx = 1:numTrainFutures varargout{1}(idx) = parfeval(@helperSpecSenseGenerateSignals,3,params); end params.numWorkersVal = max(floor(p.NumWorkers*0.1),1); numValFutures = params.numWorkersVal; varargout{2}(1:numValFutures) = parallel.FevalFuture; for idx = 1:numValFutures varargout{2}(idx) = parfeval(@helperSpecSenseGenerateSignals,3,params); end else numTestFutures = p.NumWorkers; varargout{1}(1:numTestFutures) = parallel.FevalFuture; for idx = 1:numTestFutures varargout{1}(idx) = parfeval(@helperSpecSenseGenerateSignals,3,params); end end end function [dataFuture,images,masks] = helperGetDataBatch(params,dataFuture) %helperGetDataBatch Reads one batch of data from the background data queue. [completedIdx,signals,freqLimits,labels] = fetchNext(dataFuture); dataFuture(completedIdx) = parfeval(@helperSpecSenseGenerateSignals,3,params); [images,masks] = helperSpecSenseGenerateImagesMasks(params,signals,freqLimits,labels); end function helperTrainingProgress(iter,maxTrainIter,deeplabv3_object, ... verbose,plotLoss,valFreq,t) %#ok %helperTrainingProgress Prints and/or plots the training and validation loss. if verbose trainLoss = deeplabv3_object.loss_vector{iter}; outFcnLoss = deeplabv3_object.output_loss{iter}; disp(string(t)+ " - Iteration "+iter+"/"+maxTrainIter+":") fprintf(" Train diceBCE loss: %2.2f, IOU: %2.2f\n", ... trainLoss, outFcnLoss) valIter = max(iter/valFreq,1); fprintf(" Val dice loss: %.2f, val pixel accuracy: %.2f %%\n", ... deeplabv3_object.val_dice_loss{valIter}/valIter, ... deeplabv3_object.val_accuracy{valIter}) end persistent h1 h2 if plotLoss if isempty(h1) figure; ax = gca; h1 = animatedline(ax,Color="red",Marker="o"); h2 = animatedline(ax,Color="blue",Marker="*"); title("Training Progress") xlabel("Iteration") ylabel("Training loss and IOU") legend("DiceBCE loss", "IOU") grid on end addpoints(h1,iter,deeplabv3_object.loss_vector{iter}); addpoints(h2,iter,deeplabv3_object.output_loss{iter}); drawnow end end function helperDownloadFiles() %helperDownloadFiles Download pretrained model weights. targetDir = "SpectrumSenseTorchModel"; downloadFileName = "SpectrumSenseLabV3.zip"; expFileNames = ["license.txt","specsenselabv3.pth"]; downloadData{1} = struct("DownloadFile",downloadFileName, ... "ExpectedFiles",expFileNames); dstFolder = pwd; helperDownloadExtractFile(targetDir, downloadData, dstFolder) end