randomEffects
Estimates of random effects and related statistics
Syntax
Description
Examples
Load the sample data.
load carbigFit a linear mixed-effects model for miles per gallon (MPG), with fixed effects for acceleration and horsepower, and potentially correlated random effects for intercept and acceleration, grouped by the model year. First, store the data in a table.
tbl = table(Acceleration,Horsepower,Model_Year,MPG);
Fit the model.
lme = fitlme(tbl, 'MPG ~ Acceleration + Horsepower + (Acceleration|Model_Year)');Compute the BLUPs of the random-effects coefficients and display the names of the corresponding random effects.
[B,Bnames] = randomEffects(lme)
B = 26×1
3.1270
-0.2426
-1.6532
-0.0086
1.2075
-0.2179
4.4107
-0.4887
-1.3103
-0.0208
2.8029
-0.3790
0.0865
-0.1280
0.4216
⋮
Bnames=26×3 table
Group Level Name
______________ ______ ________________
{'Model_Year'} {'70'} {'(Intercept)' }
{'Model_Year'} {'70'} {'Acceleration'}
{'Model_Year'} {'71'} {'(Intercept)' }
{'Model_Year'} {'71'} {'Acceleration'}
{'Model_Year'} {'72'} {'(Intercept)' }
{'Model_Year'} {'72'} {'Acceleration'}
{'Model_Year'} {'73'} {'(Intercept)' }
{'Model_Year'} {'73'} {'Acceleration'}
{'Model_Year'} {'74'} {'(Intercept)' }
{'Model_Year'} {'74'} {'Acceleration'}
{'Model_Year'} {'75'} {'(Intercept)' }
{'Model_Year'} {'75'} {'Acceleration'}
{'Model_Year'} {'76'} {'(Intercept)' }
{'Model_Year'} {'76'} {'Acceleration'}
{'Model_Year'} {'77'} {'(Intercept)' }
{'Model_Year'} {'77'} {'Acceleration'}
⋮
Since intercept and acceleration have potentially correlated random effects, grouped by model year of the cars, randomEffects creates a separate row for intercept and acceleration at each level of the grouping variable.
Compute the covariance parameters of the random effects.
[~,~,stats] = covarianceParameters(lme)
stats=2×1 cell array
{3×7 classreg.regr.lmeutils.titleddataset}
{1×5 classreg.regr.lmeutils.titleddataset}
stats{1}ans =
Covariance Type: FullCholesky
Group Name1 Name2 Type Estimate Lower Upper
Model_Year {'(Intercept)' } {'(Intercept)' } {'std' } 3.3475 1.2862 8.7119
Model_Year {'Acceleration'} {'(Intercept)' } {'corr'} -0.87971 -0.98501 -0.29675
Model_Year {'Acceleration'} {'Acceleration'} {'std' } 0.33789 0.1825 0.62558
The correlation value suggests that random effects seem negatively correlated. Plot the random effects for intercept versus acceleration to confirm this.
plot(B(1:2:end),B(2:2:end),'r*')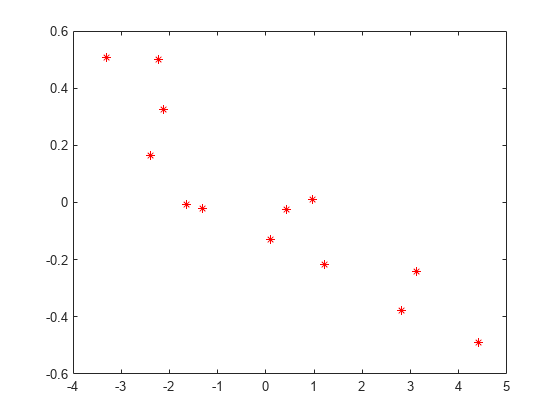
Load and display the sample data.
load fertilizer.mat
tbltbl=60×4 table
Soil Tomato Fertilizer Yield
_________ ____________ __________ _____
{'Sandy'} {'Plum' } 1 104
{'Sandy'} {'Plum' } 2 136
{'Sandy'} {'Plum' } 3 158
{'Sandy'} {'Plum' } 4 174
{'Sandy'} {'Cherry' } 1 57
{'Sandy'} {'Cherry' } 2 86
{'Sandy'} {'Cherry' } 3 89
{'Sandy'} {'Cherry' } 4 98
{'Sandy'} {'Heirloom'} 1 65
{'Sandy'} {'Heirloom'} 2 62
{'Sandy'} {'Heirloom'} 3 113
{'Sandy'} {'Heirloom'} 4 84
{'Sandy'} {'Grape' } 1 54
{'Sandy'} {'Grape' } 2 86
{'Sandy'} {'Grape' } 3 89
{'Sandy'} {'Grape' } 4 115
⋮
The table tbl contains data from a split-plot experiment, where soil is divided into three blocks based on the soil type: sandy, silty, and loamy. Each block is divided into five plots, where five different types of tomato plants (cherry, heirloom, grape, vine, and plum) are randomly assigned to these plots. The tomato plants in the plots are then divided into subplots, where each subplot is treated by one of four fertilizers. This is simulated data.
Convert Tomato, Soil, and Fertilizer to categorical variables.
tbl.Tomato = categorical(tbl.Tomato); tbl.Soil = categorical(tbl.Soil); tbl.Fertilizer = categorical(tbl.Fertilizer);
Fit a linear mixed-effects model, where Fertilizer and Tomato are the fixed-effects variables, and the mean yield varies by the block (soil type), and the plots within blocks (tomato types within soil types) independently.
lme = fitlme(tbl,"Yield ~ Fertilizer * Tomato + (1|Soil) + (1|Soil:Tomato)");Compute the BLUPs and related statistics for random effects.
[~,~,stats] = randomEffects(lme)
stats =
Random effect coefficients: DFMethod = 'Residual', Alpha = 0.05
Group Level Name Estimate SEPred tStat DF pValue Lower Upper
{'Soil' } {'Loamy' } {'(Intercept)'} 1.0061 2.3374 0.43044 40 0.66918 -3.718 5.7303
{'Soil' } {'Sandy' } {'(Intercept)'} -1.5236 2.3374 -0.65181 40 0.51825 -6.2477 3.2006
{'Soil' } {'Silty' } {'(Intercept)'} 0.51744 2.3374 0.22137 40 0.82593 -4.2067 5.2416
{'Soil:Tomato'} {'Loamy Cherry' } {'(Intercept)'} 12.46 7.1765 1.7362 40 0.090224 -2.0443 26.964
{'Soil:Tomato'} {'Loamy Grape' } {'(Intercept)'} -2.6429 7.1765 -0.36827 40 0.71461 -17.147 11.861
{'Soil:Tomato'} {'Loamy Heirloom'} {'(Intercept)'} 16.681 7.1765 2.3244 40 0.025269 2.1766 31.185
{'Soil:Tomato'} {'Loamy Plum' } {'(Intercept)'} -5.0172 7.1765 -0.69911 40 0.48853 -19.522 9.4872
{'Soil:Tomato'} {'Loamy Vine' } {'(Intercept)'} -4.6874 7.1765 -0.65316 40 0.51739 -19.192 9.8169
{'Soil:Tomato'} {'Sandy Cherry' } {'(Intercept)'} -17.393 7.1765 -2.4235 40 0.019987 -31.897 -2.8882
{'Soil:Tomato'} {'Sandy Grape' } {'(Intercept)'} -7.3679 7.1765 -1.0267 40 0.31075 -21.872 7.1364
{'Soil:Tomato'} {'Sandy Heirloom'} {'(Intercept)'} -8.621 7.1765 -1.2013 40 0.23671 -23.125 5.8833
{'Soil:Tomato'} {'Sandy Plum' } {'(Intercept)'} 7.669 7.1765 1.0686 40 0.29165 -6.8353 22.173
{'Soil:Tomato'} {'Sandy Vine' } {'(Intercept)'} 0.28246 7.1765 0.039359 40 0.9688 -14.222 14.787
{'Soil:Tomato'} {'Silty Cherry' } {'(Intercept)'} 4.9326 7.1765 0.68732 40 0.49585 -9.5718 19.437
{'Soil:Tomato'} {'Silty Grape' } {'(Intercept)'} 10.011 7.1765 1.3949 40 0.17073 -4.4935 24.515
{'Soil:Tomato'} {'Silty Heirloom'} {'(Intercept)'} -8.0599 7.1765 -1.1231 40 0.2681 -22.564 6.4444
{'Soil:Tomato'} {'Silty Plum' } {'(Intercept)'} -2.6519 7.1765 -0.36952 40 0.71369 -17.156 11.852
{'Soil:Tomato'} {'Silty Vine' } {'(Intercept)'} 4.405 7.1765 0.6138 40 0.54282 -10.099 18.909
The first three rows contain the random-effects estimates and the statistics for the three levels, Loamy, Sandy, and Silty of the grouping variable Soil. The corresponding -values 0.66918, 0.51825, and 0.82593 indicate that these random-effects are not significantly different from 0. The following 15 rows include the BLUPS of random-effects estimates for the intercept, grouped by the variable Tomato nested in Soil, i.e. interaction of Tomato and Soil.
Load the sample data.
load shiftFit a linear mixed-effects model with a random intercept grouped by operator, to assess if there is a significant difference in the performance according to the time of the shift. Use the restricted maximum likelihood method.
lme = fitlme(shift,'QCDev ~ Shift + (1|Operator)');Compute the 99% confidence intervals for random effects using the residuals option to compute the degrees of freedom. This is the default method.
[~,~,stats] = randomEffects(lme,'Alpha',0.01)stats =
Random effect coefficients: DFMethod = 'Residual', Alpha = 0.01
Group Level Name Estimate SEPred tStat DF pValue Lower Upper
{'Operator'} {'1'} {'(Intercept)'} 0.57753 0.90378 0.63902 12 0.53482 -2.1831 3.3382
{'Operator'} {'2'} {'(Intercept)'} 1.1757 0.90378 1.3009 12 0.21772 -1.5849 3.9364
{'Operator'} {'3'} {'(Intercept)'} -2.1715 0.90378 -2.4027 12 0.033352 -4.9322 0.58909
{'Operator'} {'4'} {'(Intercept)'} 2.3655 0.90378 2.6174 12 0.022494 -0.39511 5.1261
{'Operator'} {'5'} {'(Intercept)'} -1.9472 0.90378 -2.1546 12 0.052216 -4.7079 0.81337
Compute the 99% confidence intervals for random effects using the Satterthwaite approximation to compute the degrees of freedom.
[~,~,stats] = randomEffects(lme,'DFMethod','satterthwaite','Alpha',0.01)
stats =
Random effect coefficients: DFMethod = 'Satterthwaite', Alpha = 0.01
Group Level Name Estimate SEPred tStat DF pValue Lower Upper
{'Operator'} {'1'} {'(Intercept)'} 0.57753 0.90378 0.63902 6.4253 0.5449 -2.684 3.839
{'Operator'} {'2'} {'(Intercept)'} 1.1757 0.90378 1.3009 6.4253 0.23799 -2.0858 4.4372
{'Operator'} {'3'} {'(Intercept)'} -2.1715 0.90378 -2.4027 6.4253 0.050386 -5.433 1.09
{'Operator'} {'4'} {'(Intercept)'} 2.3655 0.90378 2.6174 6.4253 0.037302 -0.89598 5.627
{'Operator'} {'5'} {'(Intercept)'} -1.9472 0.90378 -2.1546 6.4253 0.071626 -5.2087 1.3142
The Satterthwaite method usually produces smaller values for the degrees of freedom (DF), which results in larger p-values (pValue) and larger confidence intervals (Lower and Upper) for the random-effects estimates.
Input Arguments
Linear mixed-effects model, specified as a LinearMixedModel object constructed using fitlme or fitlmematrix.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: [B,Bnames,stats] =
randomEffects(lme,'Alpha',0.01)
Significance level, specified as the comma-separated pair consisting of
'Alpha' and a scalar value in the range 0 to 1. For a value α,
the confidence level is 100*(1–α)%.
For example, for 99% confidence intervals, you can specify the confidence level as follows.
Example: 'Alpha',0.01
Data Types: single | double
Method for computing approximate degrees of freedom for the t-statistics
that test the random-effects coefficients against 0, specified as
the comma-separated pair consisting of 'DFMethod' and
one of the following.
"residual" | Default. The degrees of freedom are assumed to be constant and equal to n – p, where n is the number of observations and p is the number of fixed effects. |
"satterthwaite" | Satterthwaite approximation. |
"none" | All degrees of freedom are set to infinity. |
For example, you can specify the Satterthwaite approximation as follows.
Example: 'DFMethod','satterthwaite'
Output Arguments
Estimated best linear unbiased predictors of random effects
of linear mixed-effects model lme, returned as
a column vector.
Suppose lme has R grouping
variables g1, g2, ...,
gR, with levels m1, m2,
..., mR,
respectively. Also suppose q1, q2,
..., qR are
the lengths of the random-effects vectors that are associated with
g1, g2, ..., gR,
respectively. Then, B is a column vector of length q1*m1 + q2*m2 +
... + qR*mR.
randomEffects creates B by
concatenating the best linear unbiased predictors of random-effects
vectors corresponding to each level of each grouping variable as [g1level1;
g1level2; ...; g1levelm1;
g2level1; g2level2;
...; g2levelm2;
...; gRlevel1;
gRlevel2;
...; gRlevelmR]'.
Names of random-effects coefficients in B,
returned as a table.
Estimates of random effects BLUPs and related statistics, returned as a dataset array that has one row for each of the fixed effects and one column for each of the following statistics.
Group | Grouping variable associated with the random effect |
Level | Level within the grouping variable corresponding to the random effect |
Name | Name of the random-effect coefficient |
Estimate | Best linear unbiased predictor (BLUP) of random effect |
SEPred | Standard error of the estimate (BLUP minus random effect) |
tStat | t-statistic for a test that the random effect is zero |
DF | Estimated degrees of freedom for the t-statistic |
pValue | p-value for the t-statistic |
Lower | Lower limit of a 95% confidence interval for the random effect |
Upper | Upper limit of a 95% confidence interval for the random effect |
Version History
Introduced in R2013b
See Also
LinearMixedModel | fitlme | coefCI | coefTest | fixedEffects
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)