kfoldMargin
Classification margins for cross-validated linear classification model
Syntax
Description
m = kfoldMargin(CVMdl)CVMdl. That is, for every fold,
kfoldMargin estimates the classification margins for
observations that it holds out when it trains using all other observations.
m contains classification margins for each regularization strength in the
linear classification models that comprise CVMdl.
Examples
Load the NLP data set.
load nlpdataX is a sparse matrix of predictor data, and Y is a categorical vector of class labels. There are more than two classes in the data.
The models should identify whether the word counts in a web page are from the Statistics and Machine Learning Toolbox™ documentation. So, identify the labels that correspond to the Statistics and Machine Learning Toolbox™ documentation web pages.
Ystats = Y == 'stats';Cross-validate a binary, linear classification model that can identify whether the word counts in a documentation web page are from the Statistics and Machine Learning Toolbox™ documentation.
rng(1); % For reproducibility CVMdl = fitclinear(X,Ystats,'CrossVal','on');
CVMdl is a ClassificationPartitionedLinear model. By default, the software implements 10-fold cross validation. You can alter the number of folds using the 'KFold' name-value pair argument.
Estimate the cross-validated margins.
m = kfoldMargin(CVMdl); size(m)
ans = 1×2
31572 1
m is a 31572-by-1 vector. m(j) is the average of the out-of-fold margins for observation j.
Plot the k-fold margins using box plots.
figure;
boxplot(m);
h = gca;
h.YLim = [-5 30];
title('Distribution of Cross-Validated Margins')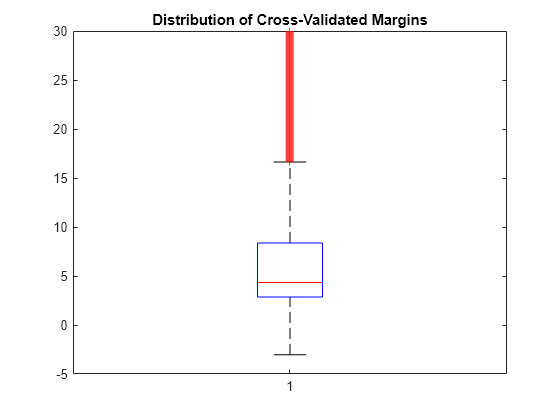
One way to perform feature selection is to compare k-fold margins from multiple models. Based solely on this criterion, the classifier with the larger margins is the better classifier.
Load the NLP data set. Preprocess the data as in Estimate k-Fold Cross-Validation Margins.
load nlpdata Ystats = Y == 'stats'; X = X';
Create these two data sets:
fullXcontains all predictors.partXcontains 1/2 of the predictors chosen at random.
rng(1); % For reproducibility p = size(X,1); % Number of predictors halfPredIdx = randsample(p,ceil(0.5*p)); fullX = X; partX = X(halfPredIdx,:);
Cross-validate two binary, linear classification models: one that uses the all of the predictors and one that uses half of the predictors. Optimize the objective function using SpaRSA, and indicate that observations correspond to columns.
CVMdl = fitclinear(fullX,Ystats,'CrossVal','on','Solver','sparsa',... 'ObservationsIn','columns'); PCVMdl = fitclinear(partX,Ystats,'CrossVal','on','Solver','sparsa',... 'ObservationsIn','columns');
CVMdl and PCVMdl are ClassificationPartitionedLinear models.
Estimate the k-fold margins for each classifier. Plot the distribution of the k-fold margins sets using box plots.
fullMargins = kfoldMargin(CVMdl); partMargins = kfoldMargin(PCVMdl); figure; boxplot([fullMargins partMargins],'Labels',... {'All Predictors','Half of the Predictors'}); h = gca; h.YLim = [-30 60]; title('Distribution of Cross-Validated Margins')
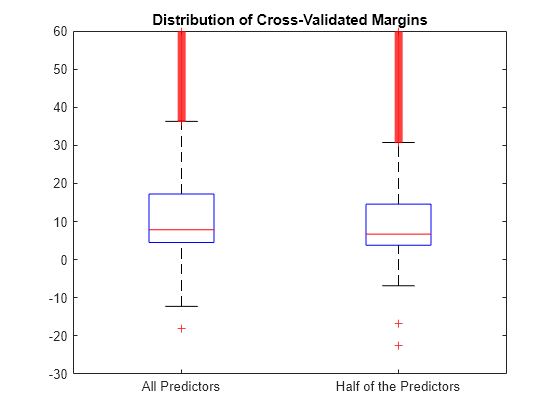
The distributions of the margins of the two classifiers are similar.
To determine a good lasso-penalty strength for a linear classification model that uses a logistic regression learner, compare distributions of k-fold margins.
Load the NLP data set. Preprocess the data as in Estimate k-Fold Cross-Validation Margins.
load nlpdata Ystats = Y == 'stats'; X = X';
Create a set of 11 logarithmically-spaced regularization strengths from through .
Lambda = logspace(-8,1,11);
Cross-validate a binary, linear classification model using 5-fold cross-validation and that uses each of the regularization strengths. Optimize the objective function using SpaRSA. Lower the tolerance on the gradient of the objective function to 1e-8.
rng(10); % For reproducibility CVMdl = fitclinear(X,Ystats,'ObservationsIn','columns','KFold',5, ... 'Learner','logistic','Solver','sparsa','Regularization','lasso', ... 'Lambda',Lambda,'GradientTolerance',1e-8)
CVMdl =
ClassificationPartitionedLinear
CrossValidatedModel: 'Linear'
ResponseName: 'Y'
NumObservations: 31572
KFold: 5
Partition: [1×1 cvpartition]
ClassNames: [0 1]
ScoreTransform: 'none'
Properties, Methods
CVMdl is a ClassificationPartitionedLinear model. Because fitclinear implements 5-fold cross-validation, CVMdl contains 5 ClassificationLinear models that the software trains on each fold.
Estimate the k-fold margins for each regularization strength.
m = kfoldMargin(CVMdl); size(m)
ans = 1×2
31572 11
m is a 31572-by-11 matrix of cross-validated margins for each observation. The columns correspond to the regularization strengths.
Plot the k-fold margins for each regularization strength. Because logistic regression scores are in [0,1], margins are in [-1,1]. Rescale the margins to help identify the regularization strength that maximizes the margins over the grid.
figure boxplot(10000.^m) ylabel('Exponentiated test-sample margins') xlabel('Lambda indices')

Several values of Lambda yield k-fold margin distributions that are compacted near 10000. Higher values of lambda lead to predictor variable sparsity, which is a good quality of a classifier.
Choose the regularization strength that occurs just before the centers of the k-fold margin distributions start decreasing.
LambdaFinal = Lambda(5);
Train a linear classification model using the entire data set and specify the desired regularization strength.
MdlFinal = fitclinear(X,Ystats,'ObservationsIn','columns', ... 'Learner','logistic','Solver','sparsa','Regularization','lasso', ... 'Lambda',LambdaFinal);
To estimate labels for new observations, pass MdlFinal and the new data to predict.
Input Arguments
Cross-validated, binary, linear classification model, specified as a ClassificationPartitionedLinear model object. You can create a
ClassificationPartitionedLinear model using fitclinear and specifying any one of the cross-validation, name-value
pair arguments, for example, CrossVal.
To obtain estimates, kfoldMargin applies the same data used to
cross-validate the linear classification model (X and Y).
Output Arguments
Cross-validated classification margins, returned as a numeric vector or matrix.
m is n-by-L, where
n is the number of observations in the data that
created CVMdl (see X and Y) and
L is the number of regularization strengths in
CVMdl (that is,
numel(CVMdl.Trained{1}.Lambda)).
m( is
the cross-validated classification margin of observation i using
the linear classification model that has regularization strength i,j)CVMdl.Trained{1}.Lambda(.j)
Data Types: single | double
More About
The classification margin for binary classification is, for each observation, the difference between the classification score for the true class and the classification score for the false class.
The software defines the classification margin for binary classification as
x is an observation. If the true label of x is the positive class, then y is 1, and –1 otherwise. f(x) is the positive-class classification score for the observation x. The classification margin is commonly defined as m = yf(x).
If the margins are on the same scale, then they serve as a classification confidence measure. Among multiple classifiers, those that yield greater margins are better.
For linear classification models, the raw classification score for classifying the observation x, a row vector, into the positive class is defined by
For the model with regularization strength j, is the estimated column vector of coefficients (the model property
Beta(:,j)) and is the estimated, scalar bias (the model property
Bias(j)).
The raw classification score for classifying x into the negative class is –f(x). The software classifies observations into the class that yields the positive score.
If the linear classification model consists of logistic regression learners, then the
software applies the 'logit' score transformation to the raw
classification scores (see ScoreTransform).
Extended Capabilities
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2016akfoldMargin fully supports GPU arrays.
Starting in R2023b, the following classification model object functions use observations with missing predictor values as part of resubstitution ("resub") and cross-validation ("kfold") computations for classification edges, losses, margins, and predictions.
In previous releases, the software omitted observations with missing predictor values from the resubstitution and cross-validation computations.
See Also
ClassificationPartitionedLinear | kfoldEdge | ClassificationLinear | kfoldPredict | margin
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)